Journal of APPLIED BIOMEDICINE
ISSN 1214-0287 (on-line)
ISSN 1214-021X (printed)
Volume 11 (2013), No 3, p 115-129
DOI 10.2478/v10136-012-0035-6
Assessing colorectal cancer heterogeneity: one step closer to tailored medicine
Pavel Pitule, Miroslava Cedikova, Vladislav Treska, Milena Kralickova, Vaclav Liska
Address: Pavel Pitule, Department of Histology and Embryology, Faculty of Medicine, Karlovarska 48, 301 00 Plzen, Czech Republic
pitulep@seznam.cz
Received 2nd January 2013.
Revised 28th January 2013.
Published online 1st February 2013.
Full text article (pdf)
Summary
Key words
Introduction
Genetic and epigenetic subtypes of CRC
Molecular markers with roles in disease prognosis and treatment prediction
Other pathways in CRC development
Non-invasive detection of CRC heterogenity
Conclusion
Support
References
SUMMARY
Many advances in understanding colorectal cancer heterogeneity and its impact on the variability of treatment efficacy have been achieved in recent years. New methods have also been introduced in colorectal cancer diagnosis and early detection, including molecular biology techniques as well as newly developed or improved imaging techniques. We are currently aware of some aspects of colorectal cancer heterogeneity, such as alterations in the epidermal growth factor receptor signalling or the different behaviours of tumours belonging to different genetic and epigenetic subtypes. In the future, greater attention should also be focused on other signalling circuits with the goal to treat patients individually, based on the characteristics of their tumours. This so-called personalised medicine will bring more benefits to patients, without unnecessary adverse side effects. Therefore, all new information regarding colorectal cancer biology brings us one step closer to accomplishing this goal.
KEY WORDS
colorectal cancer; chromosomal instability; microsatellite instability; CpG island methylator phenotype; EGFR pathway; circulating tumour cells; stool DNA
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant diseases worldwide. According to the most recent data from the GLOBOCAN project, colorectal cancer, with its more than 1.2 million new cases per year, is the third most common type of tumour and results in 600 thousand deaths per year, ranking it fourth in terms of mortality (Ferlay et al. 2010). The biggest problem is the high ability of colorectal carcinoma to form secondary tumours, particularly in the liver and lung. Based on the different studies, 20% of patients have synchronous metastasis at the time of primary tumour identification, and more than 30% of patients develop metachronous metastasis during disease progression (Mejia et al. 2012).
Another complication connected to colorectal carcinoma is the high heterogeneity of the genetic and epigenetic changes among the individual tumours. In the past, all colorectal cancers were treated as the same tumour, and the only division was based on the hereditability of this malignancy; there were hereditary and sporadic CRCs. With the development of molecular biology and spreading of its methods into clinical medicine, it has become apparent that the division of CRCs into these two groups is insufficient. Based on the major genetic and epigenetic changes, we started to recognise three main subtypes of CRC displaying different clinicopathological features (Walther et al. 2009). Recently, the complexity of to a particular chemotherapy or monoclonal antibody these subtypes was further increased by the presence treatment. Continual development of CRC is or absence of specific mutations in signalling described on the Fig. 1. pathways that can modify the response of tumours to a particular chemotherapy or monoclonal antibody treatment. Continual development of CRC is described in the Fig. 1.

Fig. 1. Continual development of colorectal cancer. Cells of the colon crypt accumulate mutations and start to proliferate. In the green arrows you can see inactivation of antioncogenes, in the red arrows are mentioned most important changes in oncogenes.
Currently, surgical intervention still has an irreplaceable role in CRC treatment because it potentially removes the entire volume of the primary or secondary tumour without respect to its molecular-biological characteristics (Mulsow et al. 2011, Kosinski et al. 2012). The subsequent oncological treatment for the eradication of micrometastatic disease or circulating tumour cells is highly variable. The most common chemotherapeutics used in CRC management are based on 5-fluorouracil (5-FU) (often in combination with leucovorin), the active derivatives of platin (oxaliplatin) and irinotecan (an inhibitor of the nuclear enzyme Topoisomerase I) (Ismaili 2011). The large spectrum of chemotherapeutics was recently enriched with the possibility of biological treatment with the monoclonal antibodies (moAb) cetuximab and panitumumab, which target and block the functioning of the epidermal growth factor receptor (EGFR), thereby stopping the signalling cascade important for the growth and division of cancer cells (Wu et al. 2008, Markman et al. 2010, Garett and Eng 2011, Kohne et al. 2012). Unfortunately, none of these treatment types are universal for all CRC patients, and treatment responses vary dramatically. Additional information regarding CRC subtype, the presence of mutations and their roles in CRC development and maintenance is therefore crucial for the identification of the best course of treatment for the individual patient and for the application of "individualised medicine".
GENETIC AND EPIGENETIC SUBTYPES OF CRC
There are three subtypes of CRC based on major genetic or epigenetic changes: tumours with chromosomal instability (CIN), those with microsatellite instability (MSI) and tumours with a CpG island methylator phenotype (CIMP) (Markowitz and Bertagnolli 2009, Perea et al. 2011, Armaghany et al. 2012). The most common subtype is CRC with chromosomal instability, which can be found in 80-85% of CRCs (Grady and Carethers 2008). MSI-positive tumours comprise 10-15% of CRCs (Malesci et al. 2007), and based on the number of altered markers, we can further distinguish the following three subtypes: MSI-high, MSI-low and microsatellite-stable (MSS). The frequency of CIMP varies between 12 to 25% of CRCs (Samowitz et al. 2005a), and we can further divide this subtype into CIMP-high and CIMP-low tumours, depending on the amount of methylated markers (Zlobec et al. 2011). The individual subtypes have different prognostic and predictive impacts on patients and are often grouped with specific mutations, which can further alter the prognosis and/ or response to the selected treatment. In addition, these subtypes are not mutually exclusive, and often CRCs present with characteristics of more than one genetic and epigenetic subtype. Relatively common combinations include MSI+ and CIMP+ (Kang 2011) or MSI+ and CIN+ (Sinicrope et al. 2006). From a clinical point of view, determination of the main subtype can be helpful for the selection of appropriate chemotherapeutic treatment and for an accurate disease prognosis (Walther et al. 2009).
Chromosomal instability
The chromosomal instability pathway is the most common mechanism leading to CRC development. It can be described as global changes in the chromosome number (aneuploidy) accompanied with a loss of heterozygosity. The loss of part or all of a chromosome leads to a physical disappearance of 25-30% of alleles (Lengauer et al. 1998). There exist several different mechanisms for CIN development, including defects in chromosomal segregation (Wang et al. 2004), centrosome abnormalities (associated with aberrant expression of the genes for Aurora and Polo-like kinases) (Ganem et al. 2009, Lassmann et al. 2009, Han et al. 2012) or telomere dysfunction (O'Hagan et al. 2002, Murnane 2006, 2012). Along with karyotypic changes, specific mutations are found in the genomes of CIN-positive CRCs. It is not clear whether these mutations are products of CIN or if CIN is the product of particular mutations. These mutations affect pathways with important roles in CRC pathogenesis. The most frequently mutated tumour suppressor genes are APC, whose protein product is a major regulator of Wnt/beta-catenin signalling and the cytoskeleton (Phelps et al. 2009), the TP53 gene, which is a regulator of transcription and cellular stress response (Zuckerman et al. 2009) and three genes located on the long arm of chromosome 18 (SMAD4, SMAD2 and DCC), which are frequently affected by allelic loss of this region (this loss is typical in greater than 70% of CRCs) (Fearon and Vogelstein 1990). Among the oncogenes, the most commonly mutated genes are CTNNB1, encoding the beta-catenin protein, which has a very important role in CRC tumourigenesis (White et al. 2012), and KRAS and PIK3CA, which both play roles in cell survival and proliferation (Samuels and Waldman 2010). The presence of these mutations on the CIN-positive background is referred to as the chromosomal instability pathway of colorectal cancer development (Tejpar and Van Cutsem 2002). Despite an enormous effort to connect individual mutations with prognostic outcomes, none are currently in use as prognostic factors in clinical practice (Pino and Chung 2010). Generally, CIN-positive tumours are associated with less favourable outcomes than are MSI-positive CRCs (Popat and Houlston 2005, Walther et al. 2008). Revealing the pathways that lead to CIN development has aided in the identification of potential chemotherapeutic targets. The benefits of blocking the function of proteins such as the Aurora and Polo-like kinases or two proteins associated with chromosomal segregation (Eg5 and CENP-E) by small-molecule inhibitors are now being examined in preclinical and early clinical trials (data collected from www.clinicaltrials.com).
Microsatellite instability
Colorectal cancer with microsatellite instability accounts for 15% of all CRCs. A major cause of MSI development is an inactivation of DNA mismatch repair mechanisms (MRR), either by mutation or downregulation of repair gene expression by promoter hypermethylation (Söreide et al. 2006). The MMR pathway is a complex system that repairs accidental changes in DNA that arise by replication mistakes, thereby maintaining the integrity of the DNA (Jun et al. 2005, Kunkel and Erie 2005, Modrich 2006, Hsieh and Yamane 2008). The most important proteins of this pathway are MLH1, PMS2, MSH2 and MSH6, whose mutations play crucial roles in the development of the hereditary form of CRC known as Lynch syndrome. Sporadic CRCs with MSI have, in most cases, epigenetically silenced MLH1 promoters (Pino and Chung 2011). The phenotype of MMR inactivation involves a change in the length of microsatellite regions, which are mono-, di- or tri- nucleotide repetitions found in many genes. Inactivation of MMR leads to somatic mutations in the genes containing microsatellite regions (mostly by reading frame shifts and the production of shortened or non-functional proteins). Some of these genes, such as PTEN, BAX and TGFbetaRII, are important for CRC development (Iacopetta et al. 2010).
Difficulties in the determination of MSI status lie in the identification of the best markers for assessing the microsatellite region length differences. The original marker panel, approved in 1997, contained two mononucleotide repetitions (BAT25 and BAT26) and three dinucleotide repetitions (D2S123, D17S250 and D5S345) (Boland et al. 1998). However, the presence of the dinucleotide repetitions in the panel led to a misclassification of some microsatellitestable tumours as tumours with low-level MSI (MSI-L) (Murphy et al. 2006). In recent years, a new panel with five mononucleotide repetitions (BAT25, BAT26, NR21, NR22 and NR24) (Suraweera et al. 2002) has become preferred to the previous one.
From a clinical perspective, CRCs with MSI have a different phenotype than other subtypes, with a higher amount of tumour-infiltrating lymphocytes, a tendency to arise mainly in the proximal part of the large intestine and a lower differentiation status (Boland and Goel 2010). MSI status is not currently used for disease prognosis or prediction, but based on available experimental data, patients with MSI-positive tumours show better survival than patients with MSI-negative or CIN-positive tumours. This effect is further altered by the presence of other mutations in the genome (Popat and Houlston 2005). For example, a BRAF mutation on an MSI-negative background is prognostically very negative (Pai et al. 2012). MSI status is not an independent predictive marker because the results of studies are affected by the simultaneous occurrence of CIN or CIMP positivity. The effect of MSI status on treatment with 5-FU, irinotecan and oxaliplatin has been examined. In the case of 5-FU, a functional MMR system is necessary to achieve cell cycle arrest after the incorporation of 5-FU into the DNA and subsequent incorrect base pairing (Jo and Carethers 2006). In some cases, detrimental effects were reported when 5-FU treatment was used in MSI-positive patients in stage II and III of the disease (Sargent et al. 2010). In another study, patients with MSI-positive tumours in stage IV showed benefits and prolonged survival after 5-FU and leucovorin treatment (Liang et al. 2002). There are also sporadic reports indicating a higher sensitivity of MSI-high tumour cells to treatment by irinotecan (Fallik et al. 2003, Vilar et al. 2008). Recent meta-analysis did not show an association between MSI status and adjuvant chemotherapy (Des Guetz et al. 2009).
The presence of MSI can be used as an important marker for the screening of Lynch syndrome, particularly in younger patients with CRC or in families with a known genetic burden. In the case of early detection of MSI associated with germinal mutations in the MMR genes, special care and attention can be focused on these patients (Schofield et al. 2009).
CpG island methylator phenotype
A characteristic feature of the third CRC subtype is the presence of a CpG island methylator phenotype resulting from the aberrant methylation of DNA CpG islands (Toyota et al. 1999, Issa 2004). CpG islands are regions rich in cytosine and guanine that are situated in the promoter region or first exon of 70% of human genes (Saxonov et al. 2006). Normally, most of these islands are not methylated (in contrast to the CpG dinucleotides outside of promoter regions), except for those connected to imprinted genes or genes located on the inactivated X chromosome (Reik and Lewis 2005, Cotton et al. 2011). Approximately 5% of genes have aberrant methylation of CpG islands in colorectal carcinoma compared to normal tissue (Schuebel et al. 2007), which is a much greater amount than the number of genes affected by mutations (Wood et al. 2007).
Currently, there is no standard set of promoter regions for the assessment of CIMP status. Among the utilised panels for methylation level measurement, two are based on five studied regions (Chan et al. 2002, Weisenberger et al. 2006) and one is based on eight different chromosomal areas (Ogino et al. 2007). These panels have different sensitivities and specificities and results obtained by the different panels are not comparable between each other. Additionally, interpretation of the results is not unified; in addition to simple division of the tumours as CIMP-positive or CIMP-negative (Weisenberger et al. 2006), it is possible to divide them into three classes (CIMPhigh, CIMP-low and CIMP-negative - Shen et al. 2007) or four classes (CIMP-high, CIMP-low and two CIMP-negative groups depending on the TP53 mutation status) (Hinoue et al. 2012), depending on the amount of methylated promoters.
CIMP is often connected to MSI because the majority of sporadic MSI-positive tumours arise via the epigenetic silencing of MLH1 gene expression, one of the most important proteins in the MMR pathway. Combining CIMP and MSI status, we can divide CRC into 4 to 6 groups with different clinical behaviours (Ogino and Goel 2008, Kang 2011). For example, the subgroup with the CIMP-high/MSIpositive phenotype usually has disease localised in the proximal part of colon and is more common in women and older patients (Kim et al. 2009, Bae et al. 2011, Hughes et al. 2012). Another important feature is the connection of CIMP and specific mutations.
CIMP-high CRCs correlate with mutations in BRAF (Weisenberg 2006), while the previously described connection between KRAS mutation and a CIMP-low phenotype is currently controversial (Ang et al. 2010). The most serious prognosis has been observed in patients with a CIMP-high/MSI-negative phenotype combined with the BRAF mutation (Lee et al. 2008).
The role of CIMP in treatment response prediction is unclear. Some studies have described a correlation between a CIMP-high status and a benefit from 5-FU adjuvant chemotherapy (Iacopetta et al. 2008, Min et al. 2011), but this result has not been confirmed by other groups (Jover et al. 2011) and requires more research.
MOLECULAR MARKERS WITH ROLES IN DISEASE PROGNOSIS AND TREATMENT PREDICTION
In addition to the described subtypes of CRC that alter the genotype and phenotype of cells globally, there is a large number of mutations in individual genes that can largely affect disease prognosis and prediction of treatment efficacy. Despite an enormous effort to identify new prognostic and predictive markers that can start a new era of personalised medicine for CRC patients, in current clinical practice, only two genes are monitored, which modify the usage of monoclonal antibodies blocking the function of EGFR. The studied markers indicate expression level status of the receptor itself and mutation status of its proximal effector KRAS. Other genes whose functional status should help with more precise disease prediction and more effective use of treatments are in various phases of experimental research. Some of them are described in the following text.
Epidermal growth factor signalling pathways
EGFR is a receptor-tyrosine kinase belonging to the HER-Erb2 protein family (Warren and Landgraf 2006). This single transmembrane glycoprotein can be specifically activated by the binding of its cognate ligand to the extracellular domain of the receptor. After ligand binding, the receptor dimerises (there can be homodimerisation as well as the formation of heterodimers with other members of its family). Dimerisation leads to activation of the intracellular kinase domain, which phosphorylates proximal members of signalling pathways emanating from the EGFR receptor. The most important proximal effector is the KRAS protein. Among the pathways activated by EGFR signalling are the MAPK, PI3K/AKT and Jak2/Stat3 pathways, all of which have crucial roles in maintaining cellular homeostasis (Yarden and Sliwkovski 2001, Lurje and Lenz 2009) (Fig. 2). Aberrant cell proliferation and its consequences are one possible outcome of the dysregulation of these pathways (Spano et al. 2005). The role of EGFR in CRC development seems to be essential, but the physiological function of EGFR signalling in the intestinal epithelium is still a matter of debate. According to current information, EGFR signalling plays an important role in the maintenance of the intestinal stem cell population (Sato et al. 2009, 2011, Xu et al. 2011, Nautiyal et al. 2012).
The EGFR pathway has a unique position in CRC treatment because it is one of the two pathways where the indication of the targeted treatment has been approved for patients with metastatic CRC (Cunningham et al. 2004). There are two types of EGFR inhibitors - small molecule tyrosine kinase inhibitors (TKI), such as erlotinib and gefitinib, and monoclonal antibodies, such as cetuximab and panitumumab (Ng and Zhu 2008). TKIs function to block the intracellular kinase domain (Takeuchi and Ito 2001), while antibodies target the extracellular domain and block receptor dimerisation (Okamoto 2010). When this type of treatment was introduced in clinical practice, only a small percentage of treated patients (approximately 10%) benefited from monoclonal antibody usage (Cunningham et al. 2004). Further research revealed that cellular EGFR expression on its own is not sufficient for the prediction of a treatment benefit. The first marker discovered to be responsible for a lack of treatment benefit in response to the anti-EGFR monoclonal antibodies was the mutated KRAS protein. Mutations resulted in its constitutive activity and consequent independence from EGFR stimulation (Lievre et al. 2006).
KRAS mutations can be found in 40% of CRC patients (Amado et al. 2008). The KRAS mutations with the greatest impact are located in codons 12 and 13 in the second exon. These two changes impair the intrinsic hydrolytic activity of the KRAS protein and stop the degradation of GTP to GDP. The role of the KRAS mutation was first observed in small retrospective studies (De Roock et al. 2008, Lievre et al. 2008) and later confirmed in large randomised prospective studies including CRYSTAL (Cetuximab Combined with Irinotecan in First-line Therapy for Metastatic Colorectal Cancer; Van Cutsem et al. 2009), OPUS (Oxaliplatin and Cetuximab in First-line Treatment of Metastatic Colorectal Cancer; Bokemeyer et al. 2009) and PRIME (Panitumumab Randomized Trial in Combination with Chemotherapy for Metastatic Colorectal Cancer to Determine Efficacy; Douillard et al. 2010). Based on their conclusions, a benefit from moAb treatment was detected only in CRC patients with wild-type KRAS protein. This inference was essential because it allowed the stratification of patients based on the presence/absence of the mutation and protected a population of the patients (those with mutated KRAS) from the unnecessary side effects of an ineffective therapy. Several recent reports have disrupted part of this theory, as different authors have shown that not only the mutation of KRAS in general is important for treatment effect prediction, but the particular mutation type must also be assessed. For example, patients with a specific change in codon 13 (p.G13D) had a partial response to the cetuximab antibody, which was unlikely to occur in patients with mutations in other portions of the KRAS protein (De Roock et al. 2010b, Tejpar et al. 2012).
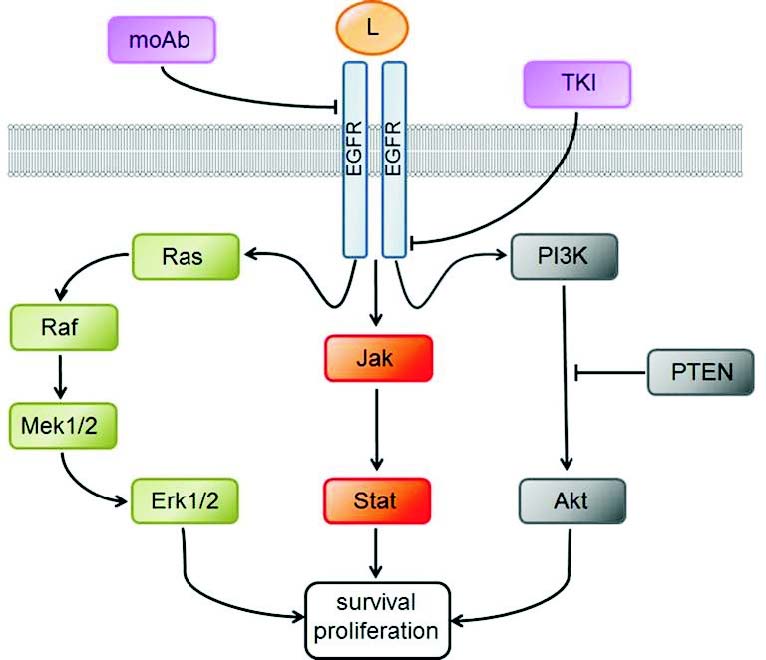
Fig. 2. Overview of the EGfR signaling pathways. The role of EGFR signaling in colorectal cancer is crucial mainly from the therapeutic point of view. EGFR function can be blocked by the monoclonal antibodies (moAb) affecting the extracellular domain by inhibiting the ligand binding (L), or by the small tyrosine kinase inhibitors (TKI), affecting the kinase domain. Three depicted pathways emanating from the EGFR are often modified in the tumour, mainly by the acquisition of different genetic/ epigenetic abberations and resulting change in the signaling capacity.
Unfortunately, even KRAS wild-type patients fail to exhibit a homogeneous response to moAb therapy. With the KRAS-mutated patients excluded, a positive reaction was still only achieved in 20-40% of patients (Vecchione et al. 2011). One possible explanation might be the functioning of another RAS family protein, NRAS. Nevertheless, it can be responsible only for a minor part of the unresponsiveness because it is mutated in only 3% of patients (De Roock et al. 2010a). Examination of other members of the EGFR signalling cascade uncovered molecular changes in additional genes that cause tumour resistance to moAb therapy, particularly mutations in the genes coding for the BRAF and PI3K proteins and decreases in PTEN expression levels (PTEN is the major negative regulator of the PI3K pathway) (Grossmann and Samowitz 2011).
BRAF is a member of the RAF kinase family and the first kinase in the MAP kinase pathway (Chong et al. 2003). This protein is mutated in 10-15% of CRC patients. The point mutation V600E causes constitutive activity of BRAF’s kinase domain (Davies et al. 2002, Samowitz et al. 2005b). Like the KRAS mutation, mutated BRAF has been repeatedly connected to tumour resistance to anti-EGFR moAb treatment (Di Nicolantonio et al. 2008, Loupakis et al. 2009). The large randomised studies OPUS and CRYSTAL did not reach a clear conclusion regarding the predictive role of BRAF mutation in the treatment with cetuximab (Bokemeyer et al. 2012). However, they were able to demonstrate a prognostic effect of BRAF mutation, as the patients with the mutated variant had shorter lengths of overall survival than the patients with wild-type BRAF (Roth et al. 2010, Yokota et al. 2011, Bokemeyer et al. 2012). Because KRAS and BRAF are both members of one signalling pathway, their mutations are mutually exclusive. For tumour program activation in the cell, it is therefore likely that only one mutation in the given signalling pathway is necessary.
A second branch of EGFR signalling is the signalling cascade commenced by phosphatidilinositol-3 kinase. This pathway, with the protein kinase AKT as the main node, performs a broad range of functions in the cell, including the regulation of glucose metabolism, gene expression, antiapoptotic actions and others (Vivanco and Sawyers 2002). Generally, this pathway regulates cell survival and metabolism. There are two changes in this pathway that have been connected to CRC development: mutation in the catalytic subunit of PI3K and change in the expression of the negative regulator protein PTEN. PIK3CA gene mutations (encoding the p100alpha catalytic subunit of PI3K) can be found in 15-25% of CRC patients (Samuels et al. 2005, De Roock et al. 2010a) and are located in exons 9 and 20 (approximately 70% of mutations are in exon 9 and 30% are in exon 20). The results of several studies focused on the predictive role of PIK3CA mutations in relation to the anti-EGFR treatment are ambiguous (Perrone et al. 2009, Prenen et al. 2009, Sartore-Bianchi et al. 2009), and it seems that a predictive role for these mutations is dependent upon the presence of other mutated genes (mainly KRAS and BRAF). Similar to the KRAS gene, each type of PIK3CA mutation has a different biological effect (De Roock et al. 2010a, Mao et al. 2012). In the case of the PTEN protein, the lack of standardised methods have caused diverse results in the individual studies, however, a connection between the loss of PTEN expression and resistance to the anti-EGFRtargeted drugs was observed (Colakoglu et al. 2008, Sawai et al. 2008, Sood et al. 2012). The roles of PTEN and PI3K expression and mutation status in treatment outcome prediction need to be validated in larger studies. The situation is very similar to the use of the PIK3CA or PTEN aberrations as prognostic markers, but recently published data have described potentially enhanced malignant behaviour in tumours with doubly mutated PIK3CA (Liao et al. 2012).
OTHER PATHWAYS IN CRC DEVELOPMENT
Despite the importance of the EGFR pathway in CRC development and treatment, other signalling cascades also have high potential for use in acquiring prognostic or predictive information or for targeting by CRC therapy. A currently existing method in CRC treatment is the blocking of VEGF signalling by the anti-VEGF monoclonal antibody bevacizumab. The VEGF pathway appears to be crucial for tumour angiogenesis, and its blocking proved to be efficient in a group of CRC patients (Hurwitz et al. 2004, Giantonio et al. 2007). This pathway can also be partially used for disease prognosis because particular single nucleotide polymorphisms in the VEGF gene were described to have prognostic roles (Vidauretta et al. 2010).
Other possible pathways crucial for CRC development have been identifiedin recent years thanks to technological advances such as whole-genome sequencing and exome scanning. These methods have described in higher detail the heterogenity of individual CRC cases, but have also revealed some common features. A recently published article from the authors contributing to The Cancer Genome Atlas Network focused on deep exon and whole-genome sequencing in 276 samples (Cancer Genome Atlas Network 2012). The authors found new types of alterations at all levels, starting from individual gene mutations, methylation or amplification and ending with chromosomal deletions or translocations. One of the most interesting results described the frequency of aberrations in entire signalling pathways. The most affected were the WNT, TGFbeta, RAS-MAPK, PI3K and p53 pathways. Another interesting fact was that almost all genes had changes in the MYC target genes, demonstrating the great importance of this oncogene in CRC development. Currently, there are only two pathways of those described above that can be blocked by targeted treatment against the EGFR, RAS-MAPK and PI3K pathways. Therefore, there is a clear need to develop new types of treatments to target the other pathways essential for tumour development and growth.
NON-INVASIVE DETECTION Of CRC HETEROGENITY
In the previous paragraphs, we attempted to convey that CRC heterogenity causes major complications in the treatment of individual patients. In clinical practice, the only method for assessing the subtype or major mutations of the CRC without surgical removal of the tumour is by biopsy, but this intervention brings additional stress to the patient. Therefore, in recent years, much effort has been dedicated to developing new, non-invasive methods that would enable us to determine the tumour subtype, its mutations and other characteristics, to better predict response to treatment. Two promising options are the detection of circulating or disseminated tumour cells, which are released from the tumour into the blood stream or bone marrow (Bidard et al. 2012), and the detection and analysis of cell-free DNA from stool samples (Miller and Steele 2012).
The fact that tumours release individual cells into bodily fluids was first observed by T. R. Ashworth almost 150 years ago (Sleijfer et al. 2007), and since this discovery, there have been attempts to use them for diagnostic as well as therapeutic purposes. However, it took until the last decades to transfer those ideas into practice, mainly because of the development of modern methods for cell isolation, detection and analysis.
The main complication is the low amount of circulating tumour cells (CTC) in contrast to normal blood or bone marrow cells. The first step in CTC detection is sample enrichment. The most common method is the use of positive immunoselection; other possibilities include negative immunoselection or centrifugation in a density gradient, which is based on the different physical properties of CTCs and blood cells. An overview of the individual types of CTC enrichment has been published elsewhere (Mikulova et al. 2011, Sun et al. 2011, Pantel and Alix-Panabieres 2012). Because CRC is a tumour of epithelial origin, it is possible to use characteristic epithelial markers for CTC enrichment, for example, expression of the cell surface protein EpCAM, or markers typical for individual types of tumours, because these markers are usually not present on normal blood cells, which are mesodermal in origin (Sleijfer et al. 2007). In initial CTC studies, lack of standardisation in the enrichment step caused heterogeneous and incomparable results among the individual studies. Even the type of anti-EpCAM antibody used for positive immunoselection could yield different results (Antolovic et al. 2010), and individual methods had different sensitivities, specificities and reproducibilities. This complication was circumvented by the introduction of several standardised systems, such as the CellSearchTM system, which was introduced in 2004 (Allard et al. 2004). This process consists of three steps: CTC enrichment using the anti-EpCAM antibody, subsequent staining of the sample with an anticytokeratin antibody (characteristic for epithelial tissue) and immunostaining with an anti-CD45 antibody, which is expressed by haematopoietic cells and serves to control for nonspecifically selected cells. The result of these three steps is a cell suspension highly enriched for CTCs, which can be used for further analysis. This system is currently widely used and many studies employing CellSearch for CTC quantification in different types of carcinomas have been published (Cohen et al. 2008, Thorsteinsson et al. 2011, Munzone et al. 2012). It is also the only system approved by the U.S. Food and Drug Administration as a method for the ancillary diagnosis of patients with metastatic CRC. The number of CTCs itself was found to be a prognostic marker in patients with metastatic CRC (Rahbari et al. 2010).
In addition to counting the CTCs in peripheral blood, it is also possible to assess the mutational status of the selected genes in the isolated CTCs and to uncover information about the primary tumour. This method is at the beginning of its development but has already been tried in several other types of tumours (Kirby et al. 2012, Magbanau et al. 2012, Sakaizawa et al. 2012). In CRC, it will be possible to focus, for example, on the mutational status of genes in the EGFR signalling cascade or even to identify the main genetic and epigenetic tumour subtype.
Cell-free stool DNA
Rapidly dividing epithelial tissues release large amount of dying cells into their surroundings. In the case of the epithelial tissue covering the gastrointestinal tract, these cells are released into the lumen along with their nucleic acids. DNA, in the form of smaller fragments, passes through the digestive system and is ultimately excreted with the stool. This DNA can be isolated from the stool and used for molecular biological analysis (Ahlquist 2010). The biggest challenge to this technique is the very low ratio of epithelial DNA to other DNA found in the stool, a major part of which originates from gut microbiota (usually >99%) (Klaassen et al. 2003). Technological advances of recent years have fortunately provided new methods such as BEAMing (Beads, Emulsions, Amplification, Magnetics) (Diehl et al. 2008) or DMC (digital melt curve) (Zou et al. 2009), which have very high sensitivities allowing the detection of specific mutations, even if present in only 0.1% of gene copies. Using these methods, it is possible to assess mutations in the KRAS or BRAF genes (Deng et al. 2012, Li et al. 2012). A very promising alternative to mutational screening is the identification of specific methylation in the isolated stool DNA. Panels of markers for the best identification of the individual gastrointestinal malignancies are currently under development (Elliott et al. 2013, Kisiel et al. 2012). Stool DNA analysis is beginning to be experimentally used as an alternative to the classical faecal blood test, mainly for preventive population screening and early tumour detection. In contrast to the common faecal blood test, stool DNA analysis has some disadvantages, including high cost and more complicated sample handling. However, it can also provide very useful information about the tumour, has very high sensitivity and can also identify tumours of the proximal regions of the gastrointestinal system (Ahlquist 2009).
CONCLUSION
As mentioned, many tumours are very heterogeneous and it is not an exception that morphologically identical tumours, which have the same tissue of origin, have developed by completely different pathways. For colorectal cancer, this heterogeneity is applicable with all its consequences. Rather than one homogeneous disease, colorectal cancer is many different types of tumours affecting one organ. However, these types are not strictly different from each other; they rather form a type of continuum of subtypes with many individual changes, which provides specific characteristics to each individual tumour. Recently, aspects of this tremendous heterogeneity have been revealed, and the most important ones were described in the individual chapters above, including novel diagnostic methods and the effects of molecular changes on patient survival.
SUPPORT
This publication was supported by grants IGA MZ CR 12025 and 13326, grant SVV 266 801, project CZ.1.05/2.1.00/03.0076 from the European Regional Development Fund and the Charles University Research Fund (project number P36).
REFERENCES
Ahlquist DA. Next-generation stool DNA testing: expanding the scope. Gastroenterology. 136: 2068-2073, 2009.
[CrossRef]
[PubMed]
Ahlquist DA. Molecular detection of colorectal neoplasia. Gastroenterology. 138: 2127-2139, 2010.
[CrossRef]
[PubMed]
Allard WJ, Matera J, Miller MC, Repollet M, Connelly MC, Rao C, Tibbe AG, Uhr JW, Terstappen LW. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 10: 6897-6904, 2004.
[CrossRef]
[PubMed]
Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 26: 1626-1634, 2008.
[CrossRef]
[PubMed]
Ang PW, Loh M, Liem N, Lim PL, Grieu F, Vaithilingam A, Platell C, Yong WP, Iacopetta B, Soong R. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer. 10: 227, 2010.
[CrossRef]
[PubMed]
Antolovic D, Galindo L, Carstens A, Rahbari N, Buchler MW, Weitz J, Koch M. Heterogeneous detection of circulating tumor cells in patients with colorectal cancer by immunomagnetic enrichment using different EpCAM-specific antibodies. BMC Biotechnol. 10: 35, 2010.
[CrossRef]
[PubMed]
Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res. 5: 19-27, 2012.
[PubMed]
Bae JM, Kim MJ, Kim JH, Koh JM, Cho NY, Kim TY, Kang GH. Differential clinicopathological features in microsatellite instability-positive colorectal cancers depending on CIMP status. Virchows Arch. 459: 55-63, 2011.
[CrossRef]
[PubMed]
Bidard FC, Ferrand FR, Huguet F, Hammel P, Louvet C, Malka D, Boige V, Ducreux M, Andre T, de Gramont A, Mariani P, Pierga JY. Disseminated and circulating tumor cells in gastrointestinal oncology. Crit Rev Oncol Hematol. 82: 103-115, 2012.
[CrossRef]
[PubMed]
Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, Loos AH, Zubel A et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 27: 663-671, 2009.
[CrossRef]
[PubMed]
Bokemeyer C, Cutsem EV, Rougier P, Ciardiello F, Heeger S, Schlichting M, Celik I, Kohne CH. Addition of cetuximab to chemotherapy as first-line treatment for KRAS wild-type metastatic colorectal cancer: pooled analysis of the CRYSTAL and OPUS randomised clinical trials. Eur J Cancer. 48: 1466-1475, 2012.
[CrossRef]
[PubMed]
Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 138: 2073-2087, 2010.
[CrossRef]
[PubMed]
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, Srivastava S. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 58: 5248-5257, 1998.
[PubMed]
Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 487: 330-337, 2012.
[CrossRef]
[PubMed]
Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, Picus J, Morse M, Mitchell E, Miller MC, Doyle GV, Tissing H et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 26: 3213-3221, 2008.
[CrossRef]
[PubMed]
Colakoglu T, Yildirim S, Kayaselcuk F, Nursal TZ, Ezer A, Noyan T, Karakayali H, Haberal M. Clinicopathological significance of PTEN loss and the phosphoinositide 3-kinase/Akt pathway in sporadic colorectal neoplasms: is PTEN loss predictor of local recurrence? Am J Surg. 195: 719-725, 2008.
[CrossRef]
[PubMed]
Cotton AM, Lam L, Affleck JG, Wilson IM, Pe?aherrera MS, McFadden DE, Kobor MS, Lam WL, Robinson WP, Brown CJ. Chromosome-wide DNA methylation analysis predicts human tissue-specific X inactivation. Hum Genet. 130: 187-201, 2011.
[CrossRef]
[PubMed]
Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 351: 337-345, 2004.
[CrossRef]
[PubMed]
Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E et al. Mutations of the BRAF gene in human cancer. Nature. 417: 949-954, 2002.
[CrossRef]
[PubMed]
De Roock W, Piessevaux H, De Schutter J, Janssens M, De Hertogh G, Personeni N, Biesmans B, Van Laethem JL, Peeters M, Humblet Y, Van Cutsem E, Tejpar S. KRAS wild-type state predicts survival and is associated to early radiological response in metastatic colorectal cancer treated with cetuximab. Ann Oncol. 19: 508-515, 2008.
[CrossRef]
[PubMed]
De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 11: 753-762, 2010a.
[CrossRef]
De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, Lamba S, Arena S, Frattini M, Piessevaux H, Van Cutsem E, O’Callaghan CJ et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 304: 1812-1820, 2010b.
[CrossRef]
[PubMed]
Deng L, Qi Z, Zou B, Wu H, Huang H, Kajiyama T, Kambara H, Zhou G. Digital detection of multiple minority mutants in stool DNA for noninvasive colorectal cancer diagnosis. Anal Chem. 84: 5645-5652, 2012.
[CrossRef]
[PubMed]
Des Guetz G, Uzzan B, Nicolas P, Schischmanoff O, Perret GY, Morere JF. Microsatellite instability does not predict the efficacy of chemotherapy in metastatic colorectal cancer. A systematic review and meta-analysis. Anticancer Res. 29: 1615-1620, 2009.
[PubMed]
Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectalcancer. J Clin Oncol. 26: 5705-5712, 2008.
[CrossRef]
[PubMed]
Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, Kinzler KW, Vogelstein B, Diaz LA, Jr. Circulating mutant DNA to assess tumor dynamics. Nat Med. 14: 985-990, 2008.
[CrossRef]
[PubMed]
Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I et al. Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) versus FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol. 28: 4697-4705, 2010.
[CrossRef]
[PubMed]
Elliott GO, Johnson IT, Scarll J, Dainty J, Williams EA, Garg D, Coupe A, Bradburn DM, Mathers JC, Belshaw NJ. Quantitative profiling of CpG island methylation in human stool for colorectal cancer detection. Int J Colorectal Dis. 28: 35-42, 2013.
[CrossRef]
[PubMed]
Fallik D, Borrini F, Boige V, Viguier J, Jacob S, Miquel C, Sabourin JC, Ducreux M, Praz F. Microsatellite instability is a predictive factor of the tumor response to irinotecan in patients with advanced colorectal cancer. Cancer Res. 63: 5738-5744, 2003.
[PubMed]
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 61: 759-767, 1990.
[CrossRef]
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 127: 2893-2917, 2010.
[CrossRef]
[PubMed]
Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460: 278-282, 2009.
[CrossRef]
[PubMed]
Garrett CR, Eng C. Cetuximab in the treatment of patients with colorectal cancer. Expert Opin Biol Ther. 11: 937-949, 2011.
[CrossRef]
[PubMed]
Giantonio BJ, Catalano PJ, Meropol NJ, O’Dwyer PJ, Mitchell EP, Alberts SR, Schwartz MA, Benson AB 3rd; Eastern Cooperative Oncology Group Study E3200. Bevacizumab in combination with oxaliplatin, fluorouracil, and leucovorin (FOLFOX4) for previously treated metastatic colorectal cancer: results from the Eastern Cooperative Oncology Group Study E3200. J Clin Oncol. 25: 1539-1544, 2007.
[CrossRef]
[PubMed]
Grady WM, Carethers JM. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology. 135: 1079-1099, 2008.
[CrossRef]
[PubMed]
Grossmann AH, Samowitz WS. Epidermal growth factor receptor pathway mutations and colorectal cancer therapy. Arch Pathol Lab Med. 135: 1278-1282, 2011.
[CrossRef]
[PubMed]
Han DP, Zhu QL, Cui JT, Wang PX, Qu S, Cao QF, Zong YP, Feng B, Zheng MH, Lu AG. Polo-like kinase 1 is overexpressed in colorectal cancer and participates in the migration and invasion of colorectal cancer cells. Med Sci Monit. 18: BR237-246, 2012.
[PubMed]
Hinoue T, Weisenberger DJ, Lange CP, Shen H, Byun HM, Van Den Berg D, Malik S, Pan F, Noushmehr H, van Dijk CM, Tollenaar RA, Laird PW. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res. 22: 271-282, 2012.
[CrossRef]
[PubMed]
Hsieh P, Yamane K. DNA mismatch repair: molecular mechanism, cancer, and ageing. Mech Ageing Dev. 129: 391-407, 2008.
[CrossRef]
[PubMed]
Hughes LA, Khalid-de Bakker CA, Smits KM, van den Brandt PA, Jonkers D, Ahuja N, Herman JG, Weijenberg MP, van Engeland M. The CpG island methylator phenotype in colorectal cancer: progress and problems. Biochim Biophys Acta. 1825: 77-85, 2012.
[PubMed]
Hurwitz H, Fehrenbacher L, Novotny W, Cartwright T, Hainsworth J, Heim W, Berlin J, Baron A, Griffing S, Holmgren E, Ferrara N, Fyfe G et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N Engl J Med. 350: 2335-2342, 2004.
[CrossRef]
[PubMed]
Chan AO, Issa JP, Morris JS, Hamilton SR, Rashid A. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol. 160: 529-536, 2002.
[CrossRef]
Chong H, Vikis HG, Guan KL. Mechanisms of regulating the Raf kinase family. Cell Signal. 15: 463-469, 2003.
[CrossRef]
Iacopetta B, Kawakami K, Watanabe T. Predicting clinical outcome of 5-fluorouracil-based chemotherapy for colon cancer patients: is the CpG island methylator phenotype the 5-fluorouracil-responsive subgroup? Int J Clin Oncol. 13: 498-503, 2008.
[CrossRef]
[PubMed]
Iacopetta B, Grieu F, Amanuel B. Microsatellite instability in colorectal cancer. Asia Pac J Clin Oncol. 6: 260-269, 2010.
[CrossRef]
[PubMed]
Ismaili N. Treatment of colorectal liver metastases. World J Surg Oncol. 9: 154, 2011.
[CrossRef]
[PubMed]
Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 4: 988-993, 2004.
[CrossRef]
[PubMed]
Jo WS, Carethers JM. Chemotherapeutic implications in microsatellite unstable colorectal cancer. Cancer Biomark. 2: 51-60, 2006.
[PubMed]
Jover R, Nguyen TP, Perez-Carbonell L, Zapater P, Paya A, Alenda C, Rojas E, Cubiella J, Balaguer F, Morillas JD, Clofent J, Bujanda L et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology. 140: 1174-1181, 2011.
[CrossRef]
[PubMed]
Jun SH, Kim TG, Ban C. DNA mismatch repair system. Classical and fresh roles. FEBS J. 273: 1609-1619, 2006.
[CrossRef]
[PubMed]
Kang GH. Four molecular subtypes of colorectal cancer and their precursor lesions. Arch Pathol Lab Med. 135: 698-703, 2011.
[PubMed]
Kim JH, Shin SH, Kwon HJ, Cho NY, Kang GH. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch. 455: 485-494, 2009.
[CrossRef]
[PubMed]
Kirby BJ, Jodari M, Loftus MS, Gakhar G, Pratt ED, Chanel-Vos C, Gleghorn JP, Santana SM, Liu H, Smith JP, Navarro VN, Tagawa ST et al. Functional characterization of circulating tumor cells with a prostate-cancer-specific microfluidic device. PLoS One. 7: e35976, 2012.
[CrossRef]
[PubMed]
Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, Ahlquist DA. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer. 118: 2623-2631, 2012.
[CrossRef]
[PubMed]
Klaassen CH, Jeunink MA, Prinsen CF, Ruers TJ, Tan AC, Strobbe LJ, Thunnissen FB. Quantification of human DNA in feces as a diagnostic test for the presence of colorectal cancer. Clin Chem. 49: 1185-1187, 2003.
[CrossRef]
[PubMed]
Kohne CH, Hofheinz R, Mineur L, Letocha H, Greil R, Thaler J, Fernebro E, Gamelin E, Decosta L, Karthaus M. First-line panitumumab plus irinotecan/5-fluorouracil/leucovorin treatment in patients with metastatic colorectal cancer. J Cancer Res Clin Oncol. 138: 65-72, 2012.
[CrossRef]
[PubMed]
Kosinski L, Habr-Gama A, Ludwig K, Perez R. Shifting concepts in rectal cancer management: a review of contemporary primary rectal cancer treatment strategies. CA Cancer J Clin. 62: 173-202, 2012.
[CrossRef]
[PubMed]
Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 74: 681-710, 2005.
[CrossRef]
[PubMed]
Lassmann S, Danciu M, Muller M, Weis R, Makowiec F, Schulte-Monting J, Hopt UT, Werner M. Aurora A is differentially expressed and regulated in chromosomal and microsatellite instable sporadic colorectal cancers. Mod Pathol. 22: 1385-1397, 2009.
[CrossRef]
[PubMed]
Lee S, Cho NY, Choi M, Yoo EJ, Kim JH, Kang GH. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int. 58: 104-113, 2008.
[CrossRef]
[PubMed]
Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 396: 643-649, 1998.
[CrossRef]
[PubMed]
Li BS, Wang XY, Xu AG, Ma FL, Ma QY, Li Z, Liu JH, Gan AH, Yu ZJ, Zhang XH, Jiang B. High-Resolution Melting Assay (HRMA) is a Simple and Sensitive Stool-Based DNA Test for the Detection of Mutations in Colorectal Neoplasms. Clin Colorectal Cancer. 11: 280-290, 2012.
[CrossRef]
[PubMed]
Liang JT, Huang KC, Lai HS, Lee PH, Cheng YM, Hsu HC, Cheng AL, Hsu CH, Yeh KH, Wang SM, Tang C, Chang KJ. High-frequency microsatellite instability predicts better chemosensitivity to high-dose 5-fluorouracil plus leucovorin chemotherapy for stage IV sporadic colorectal cancer after palliative bowel resection. Int J Cancer. 101: 519-525, 2002.
[CrossRef]
[PubMed]
Liao X, Morikawa T, Lochhead P, Imamura Y, Kuchiba A, Yamauchi M, Nosho K, Qian ZR, Nishihara R, Meyerhardt JA, Fuchs CS, Ogino S. Prognostic role of PIK3CA mutation in colorectal cancer: cohort study and literature review. Clin Cancer Res. 18: 2257-2268, 2012.
[CrossRef]
[PubMed]
Lievre A, Bachet JB, Le Corre D, Boige V, Landi B, Emile JF, Cote JF, Tomasic G, Penna C, Ducreux M, Rougier P, Penault-Llorca F, Laurent-Puig P. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 66: 3992-3995, 2006.
[CrossRef]
[PubMed]
Lievre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouche O, Landi B, Louvet C, Andre T, Bibeau F et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 26: 374-379, 2008.
[CrossRef]
[PubMed]
Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K et al. KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer. 101: 715-721, 2009.
[CrossRef]
[PubMed]
Lurje G, Lenz HJ. EGFR signaling and drug discovery. Oncology. 77: 400-410, 2009.
[CrossRef]
[PubMed]
Magbanua MJ, Sosa EV, Scott JH, Simko J, Collins C, Pinkel D, Ryan CJ, Park JW. Isolation and genomic analysis of circulating tumor cells from castration resistant metastatic prostate cancer. BMC Cancer. 12: 78, 2012.
[CrossRef]
[PubMed]
Malesci A, Laghi L, Bianchi P, Delconte G, Randolph A, Torri V, Carnaghi C, Doci R, Rosati R, Montorsi M, Roncalli M, Gennari L et al. Reduced likelihood of metastases in patients with microsatellite-unstable colorectal cancer. Clin Cancer Res. 13: 3831-3839, 2007.
[CrossRef]
[PubMed]
Mao C, Yang ZY, Hu XF, Chen Q, Tang JL. PIK3CA exon 20 mutations as a potential biomarker for resistance to anti-EGFR monoclonal antibodies in KRAS wild-type metastatic colorectal cancer: a systematic review and meta-analysis. Ann Oncol. 23: 1518-1525, 2012.
[CrossRef]
[PubMed]
Markman B, Javier Ramos F, Capdevila J, Tabernero J. EGFR and KRAS in colorectal cancer. Adv Clin Chem. 51: 71-119, 2010.
[CrossRef]
Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 361: 2449-2460, 2009.
[CrossRef]
[PubMed]
Mejia A, Schulz S, Hyslop T, Weinberg DS, Waldman SA. Molecular staging individualizing cancer management. J Surg Oncol. 105: 468-474, 2012.
[CrossRef]
[PubMed]
Mikulova V, Kolostova K, Zima T. Methods for detection of circulating tumour cells and their clinical value in cancer patients. Folia Biol (Praha). 57: 151-161, 2011.
[PubMed]
Miller S, Steele S. Novel molecular screening approaches in colorectal cancer. J Surg Oncol. 105: 459-467, 2012.
[CrossRef]
[PubMed]
Min BH, Bae JM, Lee EJ, Yu HS, Kim YH, Chang DK, Kim HC, Park CK, Lee SH, Kim KM, Kang GH. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer. 11: 344, 2011.
[CrossRef]
[PubMed]
Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 281: 30305-30309, 2006.
[CrossRef]
[PubMed]
Mulsow J, Merkel S, Agaimy A, Hohenberger W. Outcomes following surgery for colorectal cancer with synchronous peritoneal metastases. Br J Surg. 98: 1785-1791, 2011.
[CrossRef]
[PubMed]
Munzone E, Botteri E, Sandri MT, Esposito A, Adamoli L, Zorzino L, Sciandivasci A, Cassatella MC, Rotmensz N, Aurilio G, Curigliano G, Goldhirsch A et al. Prognostic value of circulating tumor cells according to immunohistochemically defined molecular subtypes in advanced breast cancer. Clin Breast Cancer. 12: 340-346, 2012.
[CrossRef]
[PubMed]
Murnane JP. Telomeres and chromosome instability. DNA Repair (Amst). 5: 1082-1092, 2006.
[CrossRef]
[PubMed]
Murnane JP. Telomere dysfunction and chromosome instability. Mutat Res. 730: 28-36, 2012.
[CrossRef]
[PubMed]
Murphy KM, Zhang S, Geiger T, Hafez MJ, Bacher J, Berg KD, Eshleman JR. Comparison of the microsatellite instability analysis system and the Bethesda panel for the determination of microsatellite instability in colorectal cancers. J Mol Diagn. 8: 305-311, 2006.
[CrossRef]
[PubMed]
Nautiyal J, Du J, Yu Y, Kanwar SS, Levi E, Majumdar AP. EGFR regulation of colon cancer stem-like cells during aging and in response to the colonic carcinogen dimethylhydrazine. Am J Physiol Gastrointest Liver Physiol. 302: G655-663, 2012.
[CrossRef]
[PubMed]
Ng K, Zhu AX. Targeting the epidermal growth factor receptor in metastatic colorectal cancer. Crit Rev Oncol Hematol. 65: 8-20, 2008.
[CrossRef]
[PubMed]
Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 10: 13-27, 2008.
[CrossRef]
[PubMed]
Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 9: 305-314, 2007.
[CrossRef]
[PubMed]
O’Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2: 149-155, 2002.
[CrossRef]
Okamoto I. Epidermal growth factor receptor in relation to tumor development: EGFR-targeted anticancer therapy. FEBS J. 277: 309-315, 2010.
[CrossRef]
[PubMed]
Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, Arber DA, Balise RR, Tubbs RR, Shadrach B, Pai RK. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 36: 744-752, 2012.
[CrossRef]
[PubMed]
Pantel K, Alix-Panabieres C. Detection methods of circulating tumor cells. J Thorac Dis. 4: 446-447, 2012.
[PubMed]
Perea J, Lomas M, Hidalgo M. Molecular basis of colorrectal cancer: towards an individualized management? Rev Esp Enferm Dig. 103: 29-35, 2011.
[CrossRef]
[PubMed]
Perrone F, Lampis A, Orsenigo M, Di Bartolomeo M, Gevorgyan A, Losa M, Frattini M, Riva C, Andreola S, Bajetta E, Bertario L, Leo E et al. PI3KCA/PTEN deregulation contributes to impaired responses to cetuximab in metastatic colorectal cancer patients. Ann Oncol. 20: 84-90, 2009.
[CrossRef]
[PubMed]
Phelps RA, Broadbent TJ, Stafforini DM, Jones DA. New perspectives on APC control of cell fate and proliferation in colorectal cancer. Cell Cycle. 8: 2549-2556, 2009.
[CrossRef]
[PubMed]
Pino MS, Chung DC. The chromosomal instability pathway in colon cancer. Gastroenterology. 138: 2059-2072, 2010.
[CrossRef]
[PubMed]
Pino MS, Chung DC. Microsatellite instability in the management of colorectal cancer. Expert Rev Gastroenterol Hepatol. 5: 385-399, 2011.
[CrossRef]
[PubMed]
Popat S, Houlston RS. A systematic review and meta-analysis of the relationship between chromosome 18q genotype, DCC status and colorectal cancer prognosis. Eur J Cancer. 41: 2060-2070, 2005.
[CrossRef]
[PubMed]
Prenen H, De Schutter J, Jacobs B, De Roock W, Biesmans B, Claes B, Lambrechts D, Van Cutsem E, Tejpar S. PIK3CA mutations are not a major determinant of resistance to the epidermal growth factor receptor inhibitor cetuximab in metastatic colorectal cancer. Clin Cancer Res. 15: 3184-3188, 2009.
[CrossRef]
[PubMed]
Rahbari NN, Aigner M, Thorlund K, Mollberg N, Motschall E, Jensen K, Diener MK, Buchler MW, Koch M, Weitz J. Meta-analysis shows that detection of circulating tumor cells indicates poor prognosis in patients with colorectal cancer. Gastroenterology. 138: 1714-1726, 2010.
[CrossRef]
[PubMed]
Reik W, Lewis A. Co-evolution of X-chromosome inactivation and imprinting in mammals. Nat Rev Genet. 6: 403-410, 2005.
[CrossRef]
[PubMed]
Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol. 28: 466-474, 2010.
[CrossRef]
[PubMed]
Sakaizawa K, Goto Y, Kiniwa Y, Uchiyama A, Harada K, Shimada S, Saida T, Ferrone S, Takata M, Uhara H, Okuyama R. Mutation analysis of BRAF and KIT in circulating melanoma cells at the single cell level. Br J Cancer. 106: 939-946, 2012.
[CrossRef]
[PubMed]
Samowitz WS, Albertsen H, Herrick J, Levin TR, Sweeney C, Murtaugh MA, Wolff RK, Slattery ML. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 129: 837-845, 2005a.
[CrossRef]
[PubMed]
Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, Wolff RK, Slattery ML. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 65: 6063-6069, 2005b.
[CrossRef]
[PubMed]
Samuels Y, Waldman T. Oncogenic mutations of PIK3CA in human cancers. Curr Top Microbiol Immunol. 347: 21-41, 2010.
[CrossRef]
[PubMed]
Samuels Y, Diaz LA, Jr., Schmidt-Kittler O, Cummins JM, Delong L, Cheong I, Rago C, Huso DL, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell. 7: 561-573, 2005.
[CrossRef]
[PubMed]
Sargent DJ, Marsoni S, Monges G, Thibodeau SN, Labianca R, Hamilton SR, French AJ, Kabat B, Foster NR, Torri V, Ribic C, Grothey A et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J Clin Oncol. 28: 3219-3226, 2010.
[CrossRef]
[PubMed]
Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 69: 1851-1857, 2009.
[CrossRef]
[PubMed]
Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 459: 262-265, 2009.
[CrossRef]
[PubMed]
Sato T, van Es JH, Snippert HJ, Stange DE, Vries RG, van den Born M, Barker N, Shroyer NF, van de Wetering M, Clevers H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature. 469: 415-418, 2011.
[CrossRef]
[PubMed]
Sawai H, Yasuda A, Ochi N, Ma J, Matsuo Y, Wakasugi T, Takahashi H, Funahashi H, Sato M, Takeyama H. Loss of PTEN expression is associated with colorectal cancer liver metastasis and poor patient survival. BMC Gastroenterol. 8: 56, 2008.
[CrossRef]
[PubMed]
Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 103: 1412-1417, 2006.
[CrossRef]
[PubMed]
Schofield L, Watson N, Grieu F, Li WQ, Zeps N, Harvey J, Stewart C, Abdo M, Goldblatt J, Iacopetta B. Population-based detection of Lynch syndrome in young colorectal cancer patients using microsatellite instability as the initial test. Int J Cancer. 124: 1097-1102, 2009.
[CrossRef]
[PubMed]
Schuebel KE, Chen W, Cope L, Glockner SC, Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den Bosch S, van Engeland M, Ting AH et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 3: 1709-1723, 2007.
[CrossRef]
[PubMed]
Shen L, Toyota M, Kondo Y, Lin E, Zhang L, Guo Y, Hernandez NS, Chen X, Ahmed S, Konishi K, Hamilton SR, Issa JP. Integrated genetic and epigenetic analysis identifies three different subclasses of colon cancer. Proc Natl Acad Sci USA. 104: 18654-18659, 2007.
[CrossRef]
[PubMed]
Sinicrope FA, Rego RL, Halling KC, Foster N, Sargent DJ, La Plant B, French AJ, Laurie JA, Goldberg RM, Thibodeau SN, Witzig TE. Prognostic impact of microsatellite instability and DNA ploidy in human colon carcinoma patients. Gastroenterology. 131: 729-737, 2006.
[CrossRef]
[PubMed]
Sleijfer S, Gratama JW, Sieuwerts AM, Kraan J, Martens JW, Foekens JA. Circulating tumour cell detection on its way to routine diagnostic implementation? Eur J Cancer. 43: 2645-2650, 2007.
[CrossRef]
[PubMed]
Sood A, McClain D, Maitra R, Basu-Mallick A, Seetharam R, Kaubisch A, Rajdev L, Mariadason JM, Tanaka K, Goel S. PTEN gene expression and mutations in the PIK3CA gene as predictors of clinical benefit to anti-epidermal growth factor receptor antibody therapy in patients with KRAS wild-type metastatic colorectal cancer. Clin Colorectal Cancer. 11: 143-150, 2012.
[CrossRef]
[PubMed]
Soreide K, Janssen EA, Soiland H, Korner H, Baak JP. Microsatellite instability in colorectal cancer. Br J Surg. 93: 395-406, 2006.
[CrossRef]
[PubMed]
Spano JP, Fagard R, Soria JC, Rixe O, Khayat D, Milano G. Epidermal growth factor receptor signaling in colorectal cancer: preclinical data and therapeutic perspectives. Ann Oncol. 16: 189-194, 2005.
[CrossRef]
[PubMed]
Sun YF, Yang XR, Zhou J, Qiu SJ, Fan J, Xu Y. Circulating tumor cells: advances in detection methods, biological issues, and clinical relevance. J Cancer Res Clin Oncol. 137: 1151-1173, 2011.
[CrossRef]
[PubMed]
Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 123: 1804-1811, 2002.
[CrossRef]
[PubMed]
Takeuchi K, Ito F. Receptor tyrosine kinases and targeted cancer therapeutics. Biol Pharm Bull. 34: 1774-1780, 2011.
[CrossRef]
[PubMed]
Tejpar S, Van Cutsem E. Molecular and genetic defects in colorectal tumorigenesis. Best Pract Res Clin Gastroenterol. 16: 171-185, 2002.
[CrossRef]
[PubMed]
Tejpar S, Celik I, Schlichting M, Sartorius U, Bokemeyer C, Van Cutsem E. Association of KRAS G13D Tumor Mutations With Outcome in Patients With Metastatic Colorectal Cancer Treated With First-Line Chemotherapy With or Without Cetuximab. J Clin Oncol. 30: 3570-3577, 2012.
[CrossRef]
[PubMed]
Thorsteinsson M, Soletormos G, Jess P. Low number of detectable circulating tumor cells in non-metastatic colon cancer. Anticancer Res. 31: 613-617, 2011.
[PubMed]
Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA. 96: 8681-8686, 1999.
[CrossRef]
[PubMed]
Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’Haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 360: 1408-1417, 2009.
[CrossRef]
[PubMed]
Vecchione L, Jacobs B, Normanno N, Ciardiello F, Tejpar S. EGFR-targeted therapy. Exp Cell Res. 317: 2765-2771, 2011.
[CrossRef]
[PubMed]
Vidaurreta M, Sanchez-Munoz R, Veganzones S, Rafael S, Gutierrez M, de-la-Orden V, Fernandez C, Arroyo M, Cerdan FJ, Maestro de las Casas ML. Vascular endothelial growth factor gene polymorphisms in patients with colorectal cancer. Rev Esp Enferm Dig. 102: 20-31, 2010.
[CrossRef]
[PubMed]
Vilar E, Scaltriti M, Balmana J, Saura C, Guzman M, Arribas J, Baselga J, Tabernero J. Microsatellite instability due to hMLH1 deficiency is associated with increased cytotoxicity to irinotecan in human colorectal cancer cell lines. Br J Cancer. 99: 1607-1612, 2008.
[CrossRef]
[PubMed]
Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2: 489-501, 2002.
[CrossRef]
[PubMed]
Walther A, Houlston R, Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 57: 941-950, 2008.
[CrossRef]
[PubMed]
Walther A, Johnstone E, Swanton C, Midgley R, Tomlinson I, Kerr D. Genetic prognostic and predictive markers in colorectal cancer. Nat Rev Cancer. 9: 489-499, 2009.
[CrossRef]
[PubMed]
Wang Z, Cummins JM, Shen D, Cahill DP, Jallepalli PV, Wang TL, Parsons DW, Traverso G, Awad M, Silliman N, Ptak J, Szabo S et al. Three classes of genes mutated in colorectal cancers with chromosomal instability. Cancer Res. 64: 2998-3001, 2004.
[CrossRef]
[PubMed]
Warren CM, Landgraf R. Signaling through ERBB receptors: multiple layers of diversity and control. Cell Signal. 18: 923-933, 2006.
[CrossRef]
[PubMed]
Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 38: 787-793, 2006.
[CrossRef]
[PubMed]
White BD, Chien AJ, Dawson DW. Dysregulation of Wnt/beta-catenin signaling in gastrointestinal cancers. Gastroenterology. 142: 219-232, 2012.
[CrossRef]
[PubMed]
Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S et al. The genomic landscapes of human breast and colorectal cancers. Science. 318: 1108-1113, 2007.
[CrossRef]
[PubMed]
Wu M, Rivkin A, Pham T. Panitumumab: human monoclonal antibody against epidermal growth factor receptors for the treatment of metastatic colorectal cancer. Clin Ther. 30: 14-30, 2008.
[CrossRef]
[PubMed]
Xu N, Wang SQ, Tan D, Gao Y, Lin G, Xi R. EGFR, Wingless and JAK/STAT signaling cooperatively maintain Drosophila intestinal stem cells. Dev Biol. 354: 31-43, 2011.
[CrossRef]
[PubMed]
Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2: 127-137, 2001.
[CrossRef]
[PubMed]
Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, Yatabe Y. BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer. 104: 856-862, 2011.
[CrossRef]
[PubMed]
Zlobec I, Bihl M, Foerster A, Rufle A, Lugli A. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol. 225: 336-343, 2011.
[CrossRef]
[PubMed]
Zou H, Taylor WR, Harrington JJ, Hussain FT, Cao X, Loprinzi CL, Levine TR, Rex DK, Ahnen D, Knigge KL, Lance P, Jiang X, Smith DI, Ahlquist DA. High detection rates of colorectal neoplasia by stool DNA testing with a novel digital melt curve assay. Gastroenterology. 136: 459-470, 2009.
[CrossRef]
[PubMed]
Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol. 219: 3-15, 2009.
[PubMed]
|
BACK
|