Journal of APPLIED BIOMEDICINE
ISSN 1214-0287 (on-line)
ISSN 1214-021X (printed)
Volume 8 (2010), No 3, p 141-150
DOI 10.2478/v10136-009-0017-5
Nucleolin, a major conserved multifunctional nucleolar phosphoprotein of proliferating cells
Francisco Javier Medina, Fernando Gonzalez-Camacho, Ana Isabel Manzano, Antonio Manrique, Raul Herranz
Address: Francisco Javier Medina, Centro de Investigaciones Biologicas (CSIC), Ramiro de Maeztu 9, 28040 Madrid, Spain
fjmedina@cib.csic.es
Received 15th March 2010.
Revised 7th May 2010.
Published online 10th May 2010.
Full text article (pdf)
Abstract in xml format
Summary
Key words
Cell proliferation, cell cycle and ribosome
biogenesis
Nucleolin purification and general
characterization
Localization in situ of nucleolin and
relation to cell proliferation
Nucleolin in mitosis
Assessment of the functional role of nucleolin in ribosome biogenesis by means of the inactivation of the nucleolin gene
Nucleolin and auxin in plants
Acknowledgement
References
SUMMARY
Nucleolin is the major nucleolar protein of animal, plant and yeast proliferating cells. It is enriched in the most soluble nuclear or nucleolar
protein extract, containing ribonucleoproteins, from which it has been purified. It has a tripartite structure in which each domain accounts for
different functions. Despite its multifunctionality, the best characterized role of nucleolin is in the primary cleavage of pre-rRNA, an early step
of ribosome biogenesis. In the nucleolus of proliferating cells, nucleolin is mostly located in the dense fibrillar component, following a
vectorial pattern, from the periphery of fibrillar centers outwards. This pattern is lost in quiescent cells in which nucleolin is present in low
levels. Nucleolin is the most phosphorylated protein of the soluble nuclear extract. It is phosphorylated by casein kinase II and CDKA, and
phosphorylation is closely associated with the role of nucleolin in proliferating cells. During mitosis, nucleolin is transported from the mother
to the daughter cell nucleolus in the form of processing particles, together with pre-rRNA precursors and other nucleolar proteins. It forms part
of prenucleolar bodies and plays a role in nucleologenesis. Recent studies on the nucleolin function, carried out on samples with inactivated
nucleolin genes (siRNA downregulated or mutants) have evidenced that nucleolin is absolutely essential for cell proliferation, for the organization
of the nucleolus and for transcription and processing of pre-rRNA. In plants, nucleolin controls the auxin responsiveness, thus being involved in
the regulation of plant development.
KEY WORDS
ribosome biogenesis; pre-rRNA processing; protein kinases; cell cycle; prenucleolar bodies; auxin
CELL PROLIFERATION, CELL CYCLE AND RIBOSOME BIOGENESIS
In highly proliferative cells, e.g. tumour cells in
animals or meristematic cells in plants, all the basic
activities of the cell (gene expression, protein
synthesis, energetic pathways, signal transduction,
etc.) are affected in different degrees by the
mechanisms driving cell growth in order to reach the
critical size capable of allowing cell division. These
functional processes take place throughout a precisely
organized cell cycle, whose progression is strictly
regulated at several checkpoints dependent on many
different signals and inputs (Mizukami 2001, Inze; and
De Veylder 2006). Cell growth requires a continuous
supply of proteins, which is necessary for building
new cellular materials. Since ribosomes are the
cellular factories in which the mRNA code is
translated into proteins, the control of ribosome
biogenesis is necessarily a key element of
proliferation control. It has been repeatedly shown
experimentally that the process of ribosome
biogenesis is highly dependent on factors regulating
cell proliferation and cell cycle progression
(Bernstein et al. 2007).
Factors stimulating cell growth and division
produce an increase in the rate of ribosome
biogenesis, which is regulated during the cell cycle,
increasing from G1 to G2, and is stopped during
mitosis (Hernandez-Verdun and Roussel 2003).
Ribosome biogenesis is morphologically expressed by
a prominent nuclear organelle, i.e., the nucleolus, and
alterations in the rate of ribosome production have
been morphologically detected as changes in the
nucleolar structure and size and in the distribution of
its subcomponents (Risueno and Medina 1986,
Saez-Vasquez and Medina 2008).
The evolution of the nucleolus during cell cycle
periods is associated with variations in nucleolar
proteins (Cerdido and Medina 1995, Klein and
Grummt 1999). These proteins, acting as targets for
factors controlling cell proliferation and cell cycle
progression, and, at the same time, as regulators of
the rate of pre-rRNA transcription and/or processing
at different levels, could be the link connecting cell
cycle and proliferation events and regulation of
ribosome biogenesis at the molecular level (Olson
1991, Medina and Gonzalez-Camacho 2003).
The positive correlation between cell proliferation
and the activity of pre-rRNA transcription and
processing also causes alterations in the size of the
nucleolus and in the distribution of the nucleolar
components, correlated with modifications in the
proliferative state of the cell. In fact, differences in
ribosome synthesizing activity are expressed as
morphologically detectable changes in the nucleolus
(Shaw and Jordan 1995, Medina et al. 2000). Since,
as indicated above, each period of the cell cycle is
associated with a particular rate of pre-rRNA
synthesis and processing, a particular structural
pattern of the nucleolus can be ascribed to each
interphase period. These structural changes could
serve as a model for the establishment of general
patterns of structure-function relationships in the
nucleolus, and these nucleolar structural patterns
could become markers for identifying cell cycle
periods (Gonzalez-Camacho and Medina 2006b).
NUCLEOLIN PURIFICATION AND GENERAL CHARACTERIZATION
The definition and characterization of the protein
population (or the proteome) of a subcellular
organelle is greatly facilitated by applying methods of
subcellular fractionation. It was proposed by our
laboratory (Gonzalez-Camacho and Medina 2006a)
that fractionation of nuclear (or nucleolar) proteins
would be carried out according to the solubility of
proteins in buffers of increasing ionic strength. This
physical criterion, accompanied in some steps by the
use of additional reagents such as detergents or
enzymes, produced fractions of functional
significance. The proposed procedure yields five
fractions, the first of them containing proteins
associated to the nuclear envelope and remnants of
the cytoskeleton; the second, which is soluble in low
ionic strength, is called "S2 fraction", and contains
ribonucleoproteins active in nuclear RNA
metabolism; after increasing ionic strength and
digesting with DNase the result is the chromatin
fraction, and finally, the fourth and fifth fractions
correspond to the nuclear matrix and are obtained,
respectively, by solubilization in high salt
concentration and in the form of the residual pellet,
only soluble in 7 M urea under sonication
(Gonzalez-Camacho and Medina 2006a)
The soluble fraction of nuclear proteins (S2) from
proliferating cells has been indeed the object of many
studies which have shown that this fraction contains
many proteins forming RNP complexes, especially
proteins actively involved in the synthesis and
processing of pre-rRNA. The major protein of this
extract, either obtained from isolated nuclei or
nucleoli, is the nucleolar protein nucleolin, formerly
called C23 (Busch et al. 1978, Bugler et al. 1982).
This finding, firstly obtained in mammalian cells, was
corroborated by our laboratory in plant meristematic
cells (De Carcer et al. 1997). Moreover, nucleolin is
the most abundant non-ribosomal protein, not only in
the soluble S2 extract, but in the nucleolus of
proliferating cells (Lapeyre et al. 1987, Martin et al.
1992).
The structure of nucleolin, as described in
animals, plants and yeast, is composed of three
domains: a) a highly charged acidic stretch at the
amino terminus with characteristic repeats; b) a
central domain containing four (animal) or two (plant
and yeast) RNA Recognition Motifs (RRMs), each of
which contains a highly conserved
RiboNucleoProtein-1 (RNP-1) octamer motif and a
less conserved RNP-2 hexamer motif, and c) a
conserved Gly- and Arg-rich carboxy-terminal
sequence, called the GAR domain (Ginisty et al.
1999).
The complex multipartite structure of nucleolin
reflects its multifunctional role: the acidic N-terminal
region, is involved in the interaction with components
of the pre-rRNA processing complex and also in the
control of rDNA transcription (Roger et al. 2003): the
central region has been implicated in RNA binding
specificity and affinity for pre-rRNA sequences
(Serin et al. 1996), and the C-terminal region or GAR
domain interacts with several ribosomal proteins
(Bouvet et al. 1998) and also binds RNA, but in a
nonspecific manner and with low affinity (Ghisolfi et
al. 1992).
The role played by nucleolin in the primary
cleavage of pre-rRNA, occurring in the 5' external
transcribed spacer (5'ETS), is particularly well known
in mammals and yeast, involving the formation of a
snoRNP complex in which the nucleolar protein
fibrillarin and several snoRNAs have been identified
(Kondo and Inouye 1992, Ginisty et al. 2000). In
plants, a similar snoRNP complex was purified and
characterized, in which the nucleolar proteins
nucleolin and fibrilarin, as well as the snoRNAs U3
and U14 were identified, and which was shown to
bind to 5'ETS RNA and accurately cleaving it at a
cleavage site mapped in vivo (Saez-Vasquez et al.
2004).
Although nucleolin has been involved essentially
in ribosome biogenesis, it has also been implicated in
a number of additional processes that take place in the
nucleus and in the cytoplasm, including RNA pol II
transcription regulation, DNA replication, mRNA
stability/translation and assembly of RNP complexes
(see Saez-Vasquez and Medina 2008).
LOCALIZATION IN SITU OF NUCLEOLIN AND RELATION TO CELL
PROLIFERATION
Nucleolin has been localized in situ at the
ultrastructural level within the nucleolus in animal
and plant cells, by using anti-nucleolin antibodies and
the immunogold method (Escande et al. 1985, Martin
et al. 1992, De Carcer et al. 1997). The adscription of
nucleolin to one or more of the different nucleolar
subcomponents has a physiological significance,
since the correlation of these structural subnucleolar
domains with the different steps of pre-rRNA
synthesis and processing is well established. In onion
proliferating cells the quantification of immunogold
labelling resulted in a preferential localization of the
protein in the dense fibrillar component (DFC)
(Martin et al. 1992, De Carcer et al. 1997). Moreover,
we examined in detail the distribution of labelling
showing that the DFC was not evenly labelled, but
particles accumulated in the neighborhoods of
fibrillar centres (FCs). Most FCs did not contain gold
particles in their interior (Fig. 1, A1 and A2). The quantitative evaluation of the labelling distribution,
by means of the Radial Distribution Function
confirmed this estimation (not shown). Therefore,
nucleolin has a vectorial distribution in the nucleolus,
from the border of FCs outwards.
In quiescent cells, cell cycle progression and
ribosome biogenesis are arrested, nucleoli do not
contain any granular component and FCs are always
large, low in number and heterogeneous in structure,
containing condensed chromatin inclusions (Risueno
and Moreno Daaz de la Espina 1979). In these cells,
we showed that the vectorial distribution of nucleolin
was lost and labelling was dispersedly located
throughout the whole nucleolar body (Fig. 1B).
Quantitative analysis confirmed this observation.
The quantification of the level of labelling showed
almost three times more labelling in proliferating than
in quiescent cells (Fig. 1C) and was indicative of the
relationship of nucleolin with cell proliferation.
Furthermore, the in situ localization is consistent
with the implication of nucleolin in transcription of
the ribosomal genes and also in the early processing
of the pre-rRNA transcript. Transcription was shown
to take place around FCs and in the transition zone
between FCs and the DFC (Martin and Medina 1991,
Shaw and Jordan 1995, De Carcer and Medina 1999);
in this zone, nucleolin has been shown to colocalize
with a protein immunologically related to the
transcription factor UBF in onion cells (De Carcer
and Medina 1999). Early pre-rRNA processing has
been described as taking place in the region of the
DFC closer to FCs (Shaw and Jordan 1995, De Cárcer
and Medina 1999), which contains high levels of
nucleolin. The presence of nucleolin in quiescent
cells, even at lower levels than those of proliferating
cells, can be related to the existence of a fraction of
the protein which is insoluble and, consequently, is
associated to the nuclear matrix (Minguez and
Moreno Diaz de la Espina 1996) (see next section). It
is not known whether the functional role of this
protein in these cells is merely structural, or
represents a transient storage form that can be
eventually activated when cellular metabolism is
resumed.
Otherwise, a correlation of increased nucleolin
expression with cell proliferation and cell cycle
progression has been directly demonstrated in peas
(Reichler et al. 2001) and also in alfalfa (Bogre et al.
1996). In onion cells, variations of the nucleolin
levels throughout the cell cycle have been
investigated, and the highest levels have been found
in G2, the period characterized by the highest rate of
pre-rRNA synthesis and processing
(Gonzalez-Camacho and Medina 2006a).
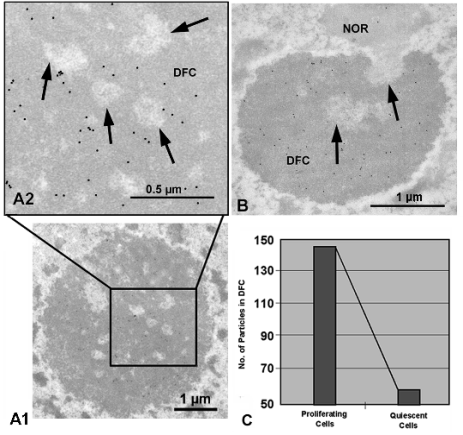
Fig. 1. Subnucleolar distribution of nucleolin after immunogold electron microscopy in onion proliferating (A1, A2) and
quiescent (B) root meristematic cells, quantitatively assessed. (A1, A2): Electron micrographs showing the ultrastructural
localization of nucleolin in proliferating cells. A2 is an enlargement of the area outlined in A1. Labelling was concentrated in
the dense fibrillar component (DFC) surrounding fibrillar centers (arrows). (B): Ultrastructural localization of nucleolin in onion
root quiescent cells. Labelling was distributed throughout the nucleolus, which is much smaller than in proliferating cells and
is exclusively formed by dense fibrillar component (DFC) and fibrillar centers (arrows), being absent the granular component.
NUCLEOLIN PHOSPHORYLATION
One of the most important features of nucleolin is
phosphorylation. Actually, the dynamics of
phosphorylation-dephosphorylation largely determine
most of the functions of the protein.
We performed a pseudo-in vivo phosphorylation
test by endogenous kinases present in the nuclei by
incubating isolated onion cell nuclei, with radioactive
orthophosphate [gamma32P]. Nuclei were then fractionated
and the S2 protein extract was electrophoretically
separated by SDS-PAGE and subjected to
autoradiography (Fig. 2A). The phosphoprotein
pattern obtained showed that the band migrating at
100 kDa, corresponding to nucleolin, was the most
phosphorylated band of the extract (Fig. 2A).
Several potential phosphorylation sites by specific
kinases have been identified in the sequences of
nucleolin both in mammals (Lapeyre et al. 1987) and
in plants (Saez-Vasquez J., personal communication).
In mammals, it was demonstrated that two kinases,
namely cdc2 kinase (or CDKA) and casein kinase II
(CKII), known to play crucial roles in the regulation
and coordination of cellular events involved in cell
cycle and proliferation, are capable of
phosphorylating nucleolin (Caizergues-Ferrer et al.
1987, Belenguer et al. 1990).
In onion cells, we tested the exogenous in vitro
phosphorylation by CKII. When it was performed on
the S2 nuclear protein extract, nucleolin (100 kDa)
was the most phosphorylated band, as visualized by
autoradiography (Fig. 2B). In addition, purified nucleolin became heavily phosphorylated in vitro by
this kinase (Fig. 2B). The additional bands appearing
in the test on purified nucleolin are due to CKII
self-phosphorylation, as shown in a control test
performed with purified CKII (Fig. 2B).
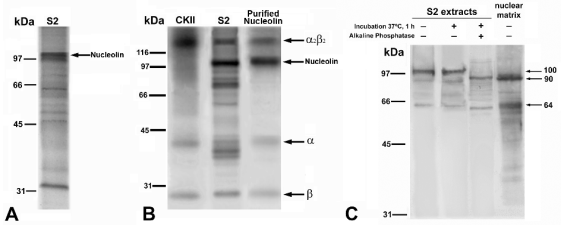
Fig. 2. Phosphorylation and dephosphorylation of nucleolin. (A): Endogenous phosphorylation of proteins of the onion root
meristematic nuclear S2 extract revealed by a pseudo-in vivo assay. Nucleolin (100 kDa) is the most phosphorylated protein. (B):
Exogenous specific phosphorylation in vitro by casein kinase II (CKII). Lane CKII: control of the self-phosphorylation of the
multimeric complex of CKII. Lane S2: phosphorylation of the S2 extract from onion root meristematic cell nuclei. Nucleolin is
the most phosphorylated protein. Lane Purified Nucleolin: in vitro phosphorylation of the purified protein. The alpha and beta subunits
of CKII are indicated. (C): In vitro dephosphorylation experiments by incubation with alkaline phosphatase of the S2 nuclear
fraction from onion meristematic cells and Western blotting with anti-nucleolin antibody. Lanes, from left to right: negative
control of the treatment, without either phosphatase or incubation; incubation without phosphatase, to show that no proteolysis
is induced by the treatment, and incubation with phosphatase, showing that the effect of dephosphorylation is a reduction in the
molecular mass of nucleolin, from 100 to 90 kDa. The lane at the right shows the result of Western blotting of the raw nuclear
matrix fraction with anti-nucleolin. The major band is 90 kDa, the same as dephosphorylated nucleolin. Molecular weight
markers of are expressed in kilodaltons (kDa).
In order to estimate the relationship of nucleolin
phosphorylation to cell proliferation, we evaluated the
phosphorylation state of nucleolin by an experiment
of in vitro dephosphorylation with alkaline
phosphatase of the S2 nuclear extract obtained from
onion root meristematic cells. Bands separated by
SDS-PAGE from either treated or untreated extracts
were visualized by Western blotting using an
anti-nucleolin antibody. Dephosphorylation resulted
in the loss of the 100 kDa band and its substitution by
a 90 kDa band, as well as in the enhancement of a
band of 64 kDa (Fig. 2C). Interestingly, these two
bands were the major bands revealed by the antibody
on intact nuclear matrix fractions obtained from the
same cells (Fig. 2C). This strongly suggests that
insoluble nucleolin, associated with the nucle(ol)ar
matrix, is in an unphosphorylated state. As mentioned
above, this insoluble form of nucleolin has a
structural role and/or is a storage form of a transitory
inactive nucleolin.
Furthermore, bi-dimensional Western blotting
with an anti-nucleolin antibody revealed 17 spots at
the level of 100 kDa in the S2 extract of meristematic
cells (Gonzalez-Camacho and Medina 2004). All the
spots detected form a cluster through a pI range of
4.3-6.6. This cluster gives an account of different
states of phosphorylation exhibited by the protein in
dependence on the nucleolar activity and the cell
cycle phases. In contrast, only 8 spots were found in
the extract from non-meristematic nuclei, whose pI
range was shortened to 4.8-6.1. This indicates a
substantially lower amount of phosphorylation
variants, associated with the drop of the proliferation
capacity and of the nucleolar activity
(Gonzalez-Camacho and Medina 2004).
NUCLEOLIN IN MITOSIS
Nucleolin is essential for the normal course of
mitosis, including chromosome congression in
metaphase (Ma et al. 2007). The localization of
nucleolin during mitosis, studied by
immunofluorescence in onion cells, showed a pair of bright spots in metaphase, presumably corresponding
to the chromosomal nucleolar organizer (NOR)
(Fig. 3A, B). In anaphase, labelling is observed at the
periphery of chromosomes, in the form of a
perichromosomal sheath. In some cases, it was
observed that the material of the perichromosomal
sheath condensed to form rounded structures,
resembling prenucleolar bodies (PNBs). Furthermore,
many small bright dots were seen in the cytoplasm,
outside the spindle (Fig. 3C, D). Finally, in telophase,
all the material revealed by the anti-nucleolin
antibody appeared either inside the reorganizing
nuclei, as PNBs very different in size to one another,
or in the cytoplasm, as small dots (Fig. 3E, F, G).
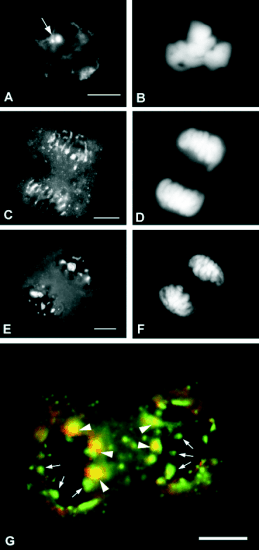
Fig. 3. Immunofluorescent localization of nucleolin
during mitosis in squashed onion root meristematic cells.
(A, C, E): nucleolin; (B, D, F): DAPI conterstaining of
DNA. (A, B): Metaphase. The nucleolar organizer (NOR;
arrow) was labelled. (C, D): Anaphase. Labelling appeared
among chromosomes, with the shape of a perichromosomal
sheath. (E, F): Telophase. Prenucleolar bodies of different
sizes were all labelled by the antibody. (G): Telophase,
double-labelled by anti-nucleolin antibody (green), and
anti-NOR90 human autoantiserum (red), detecting an
rDNA-binding protein immunologically related to the
transcription factor UBF. The picture is a merged image in
which colocalization zones appear in yellow. All
prenucleolar bodies contained nucleolin, but only a few of
them were also labelled by anti-NOR90 (arrowheads point
to colocalization; arrows point to exclusive labelling by
anti-NopA100). Prenucleolar bodies containing both
epitopes were few and larger, and they were located at
distal positions with respect to poles.
This localization of nucleolin during mitosis is
shared with other proteins involved in pre-rRNA
processing (fibrillarin, B23), and also with pre-rRNA
processing intermediates, which were synthesized in
the preceding G2. The presence of nucleolin is critical
for the assembly of these proteins and RNAs forming
the nucleolar processing complex particles called
"processomes" (Ma et al. 2007). These particles are
dispersed from the mother cell nucleolus in prophase
and are carried to the daughter cells by chromosomes
in the form of a perichromosomal sheath, which is
concentrated in PNBs at telophase, which contribute
to nucleologenesis, i.e. the formation of the new
nucleolus in daughter cells (Medina et al. 1995,
Dundr et al. 2000, Ma et al. 2007). Therefore, PNBs
are not structures which originate "de novo", but they
are formed from the materials contained in the
perichromosomal sheath which, in turn, are formed
from the dispersed remnants of the mother cell
nucleolus.
Immunolocalization of nucleolin in telophase was
combined with the detection of the antigen of the
human autoantiserum NOR90 (Fig. 3G). This serum
recognizes in mammalian cells the RNA polymerase
I transcription factor UBF (Chan et al. 1991) and it
has been shown in plant cells to recognize a
homologous epitope (Rodrigo et al. 1992),
exclusively nucleolar in localization (Rodrigo et al.
1992, De Carcer and Medina 1999). Although no
obvious UBF homolog has been found in plants, this
epitope most probably contains the HMG domain
and, therefore, has an rDNA-binding capacity.
Interestingly, the colocalization experiment revealed
two types of PNBs: those of the first type, containing
the two antigens, were few (two or three per nucleus),
larger, and appeared located at distal positions with
respect to the poles of the mother cell. The second
type of PNBs was smaller, more numerous, randomly
located in the reorganizing nucleus and was only
labelled by an anti-nucleolin antibody (Fig. 3G). The
existence of two types of PNBs, depending on
whether rDNA-bound elements of the transcription
complex are present or not, was first shown using
RNA polymerase I as the marker for the transcription
complex (Jimenez-Garcia et al. 1989). Since
pre-rRNA synthesis is resumed after mitosis in the
NOR, where the materials of PNBs, including
incompletely processed pre-rRNA molecules and
nucleolar proteins, are recruited (Dundr et al. 2000,
Medina et al. 2000) the few large UBF-containing
PNBs labelled by the anti-NOR90 serum are, in fact,
reorganizing nucleoli. The same interpretation can be
attributed to PNBs containing RNA polymerase I
(Jimenez-Garcia et al. 1989). This association of
nucleolin with rDNA in reorganizing nucleoli is
essential for nucleologenesis because
RNA-interference-mediated nucleolin-depleted cells
are unable to reorganize the nucleolus, which remains
disorganized and partially dispersed through
interphase (Ma et al. 2007).
At the time of nucleologenesis, a flow of materials
is established between PNBs and the NOR, so that
PNBs become progressively smaller whereas the new
nucleolus emerges and grows at the site of the NOR
(Savino et al. 2001). Interestingly, nucleolar protein
recruitment at the NOR occurs sequentially, the early
processing proteins being recruited first on
transcription sites, while late processing proteins
remain in PNBs for a longer time, before migrating to
the NOR (Angelier et al. 2005).
ASSESSMENT OF
THE FUNCTIONAL ROLE OF NUCLEOLIN IN RIBOSOME BIOGENESIS BY MEANS OF THE INACTIVATION OF THE NUCLEOLIN GENE
A remarkable advance regarding the function of
nucleolin comes from recent studies carried out in
both animal and plant cells using the downregulation
of the expression of the nucleolin gene, either by the
RNA-interference (siRNA)-mediated depletion, or by
mutants containing disrupted nucleolin genes. In the
preceding paragraph it was mentioned that
siRNA-downregulated nucleolin expression increased
our understanding of the role of nucleolin during
mitosis, particularly regarding the proper formation of
the metaphase plate and the nucleolus reorganization
in telophase (Ma et al. 2007).
The same approach used by another research team
on mammalian cultured cells (Ugrinova et al. 2007),
together with a more recent study based on the
generation of a nucleolin conditional knockout
(Storck et al. 2009) showed a rapid and drastic drop
of the cell proliferation rate associated with the
decrease in the levels of nucleolin; this drop was due
to the arrest of cell cycle progression, either in G1 or
in G2, accompanied by the entry in apoptosis of a
significant number of cells; furthermore, the nucleolar
substructure was re-arranged, appearing with the
components segregated, resembling the structure
shown after RNA polymerase I transcription
inhibition by actinomycin D treatment. In fact, the
loss of function of nucleolin caused a drastic
reduction in rRNA gene transcription and in contrast,
pre-rRNA processing was shown to be only slightly
affected.
In plants, the analysis of three independent
Arabidopsis nucleolin mutants, all of them
encompassing a disruption of the nucleolin gene,
showed that the lack of expression of nucleolin
phenotypically resulted in a reduced growth rate, a
prolonged life cycle, pointed leaves, a defective
vascular pattern and aberrant leaf venation (Kojima et
al. 2007, Petricka and Nelson 2007, Pontvianne et al.
2007). Therefore, nucleolin plays a major role in plant
growth and development and even has a considerable
influence on the structure of the plant. Most probably,
these effects at the level of the whole plant are the
consequence of the alteration of ribosome biogenesis,
but no direct evidence has been reported for any of
the phenotypic effects.
At the molecular level, independent analysis of the
three nucleolin mutants has consistently revealed that
the absence of the protein induces changes in
pre-rRNA processing, especially in the early cleavage
that occurs at the level of the 5'ETS (Kojima et al.
2007, Petricka and Nelson 2007, Pontvianne et al.
2007). Intermediate pre-ribosomal precursors, and
particularly the 35 S pre-rRNA (Petricka and Nelson
2007) appear to be accumulated in the mutants in
relation to the wild type. This is associated with our
finding that the nucleolus of mutant plant cells
contains the so-called nucleolar perichromatin-like
granules (Pontvianne et al. 2007), structures
characterized to as containing wrongly processed
pre-rRNA.
The molecular alterations in the nucleolin
mutants lead to nucleolar disorganization, involving
the loss of the spherical shape of the nucleolus and
the hard discrimination of nucleolar subcomponents,
being substituted by a loose accumulation of fibrils
and granules (Pontvianne et al. 2007).
Plants, in contrast to animals, contain at least two
genes encoding nucleolin or nucleolin-like proteins,
called AtNUC-L1 and AtNUC-L2 (Pontvianne et al.
2007). In Atnuc-L1 mutant plants, the homologous
AtNUC-L2 gene, which is usually repressed under
normal growing conditions, is induced to be
expressed. We have found that AtNUC-L2 localizes
in the nucleolus of Atnuc-L1 and might rescue, at
least partially, the loss of AtNUC-L1 (Pontvianne et
al. 2007).
NUCLEOLIN AND AUXIN IN PLANTS
Nucleolin loss of function in plants alters the auxin
responsiveness with regard to both auxin transport
and auxin response. It was suggested that nucleolin
might be involved in auxin-dependent control of
organ growth and patterning, potentially indicating a
correlation between auxin regulation and ribosome
biogenesis (Petricka and Nelson 2007). Eventually,
this could be seen as a way in which nucleolin
controls plant development, since the phytohormone
auxin influences multiple aspects of growth and
differentiation in plants. Low levels of auxin induce
cell elongation, enlargement and differentiation,
whereas high levels of auxin stimulate cell
proliferation and cell cycle progression (Magyar et al.
2005).
Levels of nucleolin have been found to be
depleted in plant meristematic cells grown during
paceflight, i.e. under environmental conditions of
weightlessness or microgravity; this was associated
with the uncoupling of cell growth and proliferation
in these cells (Matia et al. 2010). At the same time,
growth in microgravity results in a substantial
inhibition of the auxin polar transport (Ueda et al.
1999). This strongly suggests that the role of auxin in
the maintenance of the coupling of cell growth and
proliferation under normal gravity conditions is lost,
at least partially, as a consequence of the change in
environmental gravity (Medina and Herranz 2010).
Most likely, nucleolin plays a part in this alteration.
FUTURE PROSPECTS
Despite this intensive and extensive work, many
points remain obscure on our understanding of the
functional role of nucleolin in ribosome biogenesis
and in other cellular functions, related or not to what
has been considered up till now as the main role of
the protein; namely its participation in the synthesis
of ribosomes. Even, the highly productive and
promising method of using either mutants or siRNA
to block the expression of the nucleolin gene is still at
the beginning of its course. These are our next
challenges in the investigation of this exciting and
crucial protein.
ACKNOWLEDGEMENT
Work performed in the authors' laboratory was
supported by different Grants of the Spanish "Plan
Nacional de Investigacion Cientifica y Tecnologica",
and particularly by the Grant No.
ESP2006-13600-C02-02.
REFERENCES
Angelier N, Tramier M, Louvet E, Coppey-Moisan M, Savino TM, De Mey JR, Hernandez-Verdun D: Tracking the interactions of rRNA processing proteins
during nucleolar assembly in living cells. Mol Biol Cell 16:2862-2871, 2005.
Belenguer P, Caizergues-Ferrer M, Labbe JC, Doree M, Amalric F: Mitosis-specific phosphorylation of nucleolin by p34 cdc2 protein kinase. Mol Cell
Biol 10:3607-3618, 1990.
Bernstein KA, Bleichert F, Bean JM, Cross FR, Baserga SJ: Ribosome biogenesis is sensed at the start cell cycle checkpoint. Mol Biol Cell
18:953-964, 2007.
Bogre L, Jonak C, Mink M, Meskiene I, Traas J, Ha DTC, Swoboda I, Plank C, Wagner E, Heberle-Bors E, Hirt H: Developmental and cell cycle
regulation of alfalfa NucMs1, a plant homolog of the yeast NSR1 and mammalian nucleolin. Plant Cell 8:417-428, 1996.
Bouvet P, Diaz JJ, Kindbeiter K, Madjar JJ, Amalric F: Nucleolin interacts with several ribosomal proteins through its RGG domain. J Biol Chem
273:19025-19029, 1998.
Bugler B, Caizergues-Ferrer M, Bouche G, Bourbon HM, Amalric F: Detection and localization of a class of proteins immunologically related to a
100-kDa nucleolar protein. Eur J Biochem 128:475-480, 1982.
Busch H, Ballal NR, Rao MRS, Choi YC: Factors affecting nucleolar rDNA readouts. In Rothblum LI, Busch H (eds.): The Cell Nucleus, vol 5,
Chromatin, part b, Academic Press, New York, San Francisco, London 1978, pp. 416-468.
Caizergues-Ferrer M, Belenguer P, Lapeyre B, Amalric F, Wallace MO, Olson MOJ: Phosphorylation of nucleolin by a nuclear type NII protein kinase.
Biochemistry 26:7876-7883, 1987.
Cerdido A, Medina FJ: Subnucleolar location of fibrillarin and variation in its levels during the cell cycle and during differentiation of plant
cells. Chromosoma 103:625-634, 1995.
Chan EKL, Imai H, Hamel JC, Tan EM: Human autoantibody to RNA polymerase I transcription factor hUBF. Molecular identity of nucleolus organizer
region autoantigen NOR-90 and ribosomal RNA transcription upstream binding factor. J Exp Med 174:1239-1244, 1991.
De Carcer G, Medina FJ: Simultaneous localization of transcription and early processing markers allows dissection of functional domains in the
plant cell nucleolus. J Struct Biol 128:139-151, 1999.
De Carcer G, Cerdido A, Medina FJ: Nopa64, a novel nucleolar phosphoprotein from proliferating onion cells, sharing immunological determinants with
mammalian nucleolin. Planta 201:487-495, 1997.
Dundr M, Misteli T, Olson MOJ: The dynamics of postmitotic reassembly of the nucleolus. J Cell Biol 150:433-446, 2000.
Escande ML, Gas N, Stevens BJ: Immunolocalization of the 100 k nucleolar protein in CHO cells. Biol Cell 53:99-110, 1985.
Ghisolfi L, Joseph G, Amalric F, Erard M: The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its
RNA-helix-destabilizing properties. J Biol Chem 267:2955-2959, 1992.
Ginisty H, Sicard H, Roger B, Bouvet P: Structure and functions of nucleolin. J Cell Sci 112:761-772, 1999.
Ginisty H, Serin G, Ghisolfi-Nieto L, Roger B, Libante V, Amalric F, Bouvet P: Interaction of nucleolin with an evolutionarily conserved
pre-ribosomal RNA sequence is required for the assembly of the primary processing complex. J Biol Chem 275:18845-18850, 2000.
Gonzalez-Camacho F, Medina FJ: Identification of specific plant nucleolar phosphoproteins in a functional proteomic analysis Proteomics 4:407-417,
2004.
Gonzalez-Camacho F, Medina FJ: Extraction of nuclear proteins from root meristematic cells. In Thiellement H, Zivy M, Damerval C, Mechin V (eds.):
Plant Proteomics: Methods and Protocols, Humana Press, Totowa, NJ 2006a, pp. 63-72.
Gonzalez-Camacho F, Medina FJ: The nucleolar structure and the activity of nucleolin-like protein NopA100 during the cell cycle in proliferating
plant cells. Histochem Cell Biol 125:139-153, 2006b.
Hernandez-Verdun D, Roussel P: Regulators of nucleolar functions. Prog Cell Cycle Res 5:301-308, 2003.
Inze D, De Veylder L: Cell cycle regulation in plant development. Annu Rev Genet 40:77-105, 2006.
Jimenez-Garcia LF, Rothblum LI, Busch H, Ochs RL: Nucleologenesis:Use of non-isotopic "in situ" hybridization and immunocytochemistry to
compare the localization of rDNA and nucleolar proteins during mitosis. Biol Cell 65:239-246, 1989.
Klein J, Grummt I: Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis
and early G1. Proc Natl Acad Sci USA 96:6096-6101, 1999.
Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Kato T, Tabata S, Saez-Vasquez J, Echeverria M, Nakagawa T, Ishiguro S, Nakamura K: Sugar-inducible
expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J 49:1053-1063,
2007.
Kondo K, Inouye M: Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem
267:16252-16258, 1992.
Lapeyre B, Bourbon HM, Amalric F: Nucleolin, the major nucleolar protein of growing eukaryotic cells: An unusual protein structure revealed by the
nucleotide sequence. Proc Natl Acad Sci USA 84:1472-1476, 1987.
Ma N, Matsunaga S, Takata H, Ono-Maniwa R, Uchiyama S, Fukui K: Nucleolin functions in nucleolus formation and chromosome congression. J Cell Sci
120:2091-2105, 2007.
Magyar Z, De Veylder L, Atanassova L, Bako L, Inze D, Bogre L: The role of Arabidopsis E2FB transcription factor in regulating auxin-dependent cell
division. Plant Cell 17:2527-2541, 2005.
Martin M, Medina FJ: A Drosophila anti-RNA polymerase II antibody recognizes a plant nucleolar antigen, RNA polymerase I, which is mostly
localized in fibrillar centres. J Cell Sci 100:99-107, 1991.
Martin M, Garcia-Fernandez LF, Moreno Diaz de la Espina S, Noaillac-Depeyre J, Gas N, Medina FJ: Identification and localization of a nucleolin
homologue in onion nucleoli. Exp Cell Res 199:74-84, 1992.
Matia I, Gonzalez-Camacho F, Herranz R, Kiss JZ, Gasset G, van Loon JJWA, Marco R, Medina FJ: Plant cell proliferation and growth are altered by
microgravity conditions in spaceflight. J Plant Physiol 167:184-193 2010.
Medina FJ, Gonzalez-Camacho F: Nucleolar proteins and cell proliferation in plant cells. Recent Res Dev Plant Biol 3:55-68, 2003.
Medina FJ, Herranz R: Microgravity environment uncouples cell growth and cell proliferation in root meristematic cells: The mediator role of auxin.
Plant Signal Behav 5:176-179, 2010.
Medina FJ, Cerdido A, Fernandez-Gomez ME: Components of the nucleolar processing complex (pre-rRNA, fibrillarin and nucleolin) colocalize during
mitosis and are incorporated to daughter cell nucleoli. Exp Cell Res 221:111-125, 1995.
Medina FJ, Cerdido A, De Carcer G: The functional organization of the nucleolus in proliferating plant cells. Eur J Histochem 44:117-131,
2000.
Minguez A, Moreno Diaz de la Espina S: In situ localization of nucleolin in the plant nucleolar matrix. Exp Cell Res 222:171-178,
1996.
Mizukami Y: A matter of size: Developmental control of organ size in plants. Curr Opin Plant Biol 4:533-539, 2001.
Olson MOJ: The role of proteins in nucleolar structure and function. In Strauss PR, Wilson SH (eds.): The Eukaryotic Nucleus Molecular Biochemistry
and Macromolecular Assemblies, The Telford Press, Caldwell, New Jersey 1991, pp. 519-559.
Petricka JJ, Nelson TM: Arabidopsis nucleolin affects plant development and patterning. Plant Physiol 144:173-186, 2007.
Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J: Characterization of AtNUC-L1 reveals a central role of
nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell 18:369-379, 2007.
Reichler SA, Balk J, Brown ME, Woodruff K, Clark GB, Roux SJ: Light differentially regulates cell division and the mRNA abundance of pea nucleolin
during de-etiolation. Plant Physiol 125:339-350, 2001.
Risueno MC, Moreno Diaz de la Espina S: Ultrastructural and cytochemical study of the quiescent root meristematic cell nucleus. J Submicrosc Cytol
11:85-98, 1979.
Risueno MC, Medina FJ: The nucleolar structure in plant cells. University of the Basque Country-Springer International, Leioa 1986, 1-154
pp.
Rodrigo RM, Rendon MC, Torreblanca J, Garcia-Herdugo G, Moreno FJ: Characterization and immunolocalization of RNA polymerase I transcription factor
UBF with anti-NOR serum in Protozoa, higher plant and vertebrate cells. J Cell Sci 103:1053-1063, 1992.
Roger B, Moisand A, Amalric F, Bouvet P: Nucleolin provides a link between RNA polymerase I transcription and pre-ribosome assembly. Chromosoma
111:399-407, 2003.
Saez-Vasquez J, Medina FJ: The plant nucleolus. In Kader JC, Delseny M (eds.): Advances in Botanical Research, vol 47, Elsevier, San Diego, CA
2008, pp. 1-46.
Saez-Vasquez J, Caparros-Ruiz D, Barneche F, Echeverria M: A plant snoRNP complex containing snoRNAs, fibrillarin, and nucleolin-like proteins is
competent for both rRNA gene binding and pre-rRNA processing in vitro. Mol Cell Biol 24:7284-7297, 2004.
Savino TM, Gebrane-Younes J, De Mey J, Sibarita J-B, Hernandez-Verdun D: Nucleolar assembly of the rRNA processing machinery in living cells. J
Cell Biol 153:1097-1110, 2001.
Serin G, Joseph G, Faucher C, Ghisolfi L, Bouche G, Amalric F, Bouvet P: Localization of nucleolin binding sites on human and mouse pre- ribosomal
RNA. Biochimie 78:530-538, 1996.
Shaw PJ, Jordan EG: The nucleolus. Annu Rev Cell Biol 11:93-121, 1995.
Storck S, Thiry M, Bouvet P: Conditional knockout of nucleolin in DT40 cells reveals the functional redundancy of its RNA-binding domains. Biol
Cell 101:153-167, 2009.
Ueda J, Miyamoto K, Yuda T, Hoshino T, Fujii S, Mukai C, Kamigaichi S, Aizawa S, Yoshizaki I, Shimazu T, Fukui K: Growth and development, and auxin
polar transport in higher plants under microgravity conditions in space: Bric-aux on STS-95 space experiment. J Plant Res 112:487-492,
1999.
Ugrinova I, Monier K, Ivaldi C, Thiry M, Storck S, Mongelard F, Bouvet P: Inactivation of nucleolin leads to nucleolar disruption, cell cycle
arrest and defects in centrosome duplication. BMC Mol Biol 8:66, 2007.
|
BACK
|