Journal of APPLIED BIOMEDICINE
ISSN 1214-0287 (on-line)
ISSN 1214-021X (printed)
Volume 8 (2010), No 4, p 179-188
DOI 10.2478/v10136-009-0021-9
Bacterial toxin-antitoxin systems targeting translation
Ramon Diaz-Orejas, Elizabeth Diago-Navarro, Ana Maria Hernandez Arriaga, Juan Lopez-Villarejo, Marc Lemonnier, Inma Moreno-Cordoba, Concha Nieto, Manuel Espinosa
Address: Ramon Diaz-Orejas, Centro de Investigaciones Biologicas - CSIC, Ramiro de Maeztu 9, E-28040 Madrid, Spain
ramondiaz@cib.csic.es
Received 5th March 2010.
Revised 30th April 2010.
Published online 17th May 2010.
Full text article (pdf)
Abstract in xml format
Summary
Key words
Introduction
The parD system of plasmid R1: inhibition of protein synthesis by mRNA cleavage in the absence of ribosomes
RelBE type systems: protein inhibition by cleavage of mRNA on the ribosomes
Some possible biomedical applications/implications of TAS
Acknowledgements
References
SUMMARY
Toxin-antitoxin systems (TAS) emerged more than 25 years ago and have since developed as an important field in molecular microbiology. TAS are autoregulated operons coding a stable toxin and an unstable antitoxin found in the plasmids and chromosomes of Bacteria and Archaea. The conditional activation of their toxins interferes with cell growth/viability and, depending on the context, can influence plasmid maintenance, stress management, bacterial persistence, cell differentiation and, it is likely also bacterial virulence. This review summarizes recent results on the parD system of plasmid R1 and on the chromosomal relBE systems found in Escherichia coli and in Streptococcus pneumoniae with a focus on the RNase activity of their toxins, their regulation and their biomedical applications and implications.
KEY WORDS
toxin-antitoxin systems; plasmid maintenance; cell growth regulation; toxin RNases; protein synthesis inhibition
INTRODUCTION
Toxin-antitoxin systems (TAS) were reported more
than 25 years ago as plasmid auxiliary maintenance
cassettes. Three plasmid-borne TAS contributed
initially to establishing this field in microbiology: the
parB (hok, sok) and parD (kis, kid) systems of
plasmid R1 and the ccd (ccdA, ccdB) system of
plasmid F (Gerdes et al. 1986, Hiraga et al. 1986,
Bravo et al. 1987). TAS are based on the activity of
two intracellular components, a stable toxin and a constitutively unstable antitoxin; the differential
decay of the antitoxin, occurring also in cells losing
the plasmid at cell division, allows activation of the
toxin in these cells and its subsequent interference
with cell growth/viability. This favours the
preferential propagation of plasmid containing TAS
and contributes to their maintenance in bacterial
populations. The three systems described, initially
define two main TAS types: hok/sok is the prototype
of Type I TAS in which the toxin is a protein and the
antitoxin is an RNA, while ccd and parD define type
II TAS in which both the antitoxin and the toxin are
proteins. Most of the focus has been so far
concentrated on type II systems (Hayes 2003, Buts et
al. 2005, Gerdes et al. 2005, Condon 2006, Kamphuis
et al. 2007a, Van Melderen and Saavedra De Bast
2009).
Following the initial reports, many type II systems
were found in plasmids and also in the chromosomes
of many Eubacteria and Archaea (Pandey and Gerdes
2005). Up to 9 different type II TA families have been
defined. TA systems can contribute to stress
management in bacterial populations, to genome
stability or to complex phenotypes such as bacterial
persistence and cell differentiation and eventually as
anti-addiction modules (Van Melderen and Saavedra
De Bast 2009).
This review summarizes recent contributions
made in our two groups (RDO and MEP) on the TAS
systems parD (kis, kid) of plasmid R1 and relBE of
Streptococcus pneumoniae, whose toxins cleave
mRNAs. We focus on the mechanisms of action of
the toxins of these TAS which inhibit translation, and
on the way they are regulated. The biomedical
implications/applications of these studies is briefly
addressed.
THE parD SYSTEM OF PLASMID R1:
INHIBITION OF PROTEIN SYNTHESIS BY
mRNA CLEAVAGE IN THE ABSENCE OF
RIBOSOMES
Identification of the parD system and its specific
role in the maintenance of plasmid R1
parD (kis, kid) system was reported by our laboratory
in 1987 (Bravo et al. 1987). This TAS is a bicistronic
operon of plasmid R1 which is close to the basic
replicon of the plasmid. It codes two short proteins,
an antitoxin, Kis (killer suppressor), and a toxin, Kid (killing determinant). An identical system to parD,
pem, was also found in plasmid R100 (Tsuchimoto et
al. 1988) and two chromosomal TA systems
homologous to pem (chpA/mazEF and chpB) were
later identified by the same authors (Masuda et al.
1993). A marginal cross-talk between these systems
and parD that could be enhanced by mutations
affecting the chpAI/mazE and chpBI antitoxins could
be detected (Santos-Sierra et al. 1997, 1998). Under
standard growth conditions parD is inefficient as a
plasmid maintenance system and for this reason it
was initially undetected. It was identified by an
unexpected mutation in the kis antitoxin gene that
greatly increased the stability of the plasmid and in
combination with other mutations, revealed the toxin
and antitoxin activities of Kis and Kid (Bravo et al.
1987, 1988). The first kis mutant also indicated a key
role for the antitoxin in the transcriptional regulation
of the parD operon. Unexpectedly, the parD system
was activated when replication of a low copy mini-R1
plasmid was compromised and, surprisingly, this
activation contributed to recover the plasmid
replication (Ruiz-Echevarria et al. 1995b). This result
suggested the specialized role of the system in
recovering inefficient plasmid replication, and this
recovery was clarified following the identification of
the RNase activity of Kid (see below): it seems that
the activated Kid toxin modulates the efficiency of
replication by down-regulating the mRNA levels of
CopB, the auxiliary R1 copy number controller, via
specific RNA cleavage within the polycistronic
copB-repA mRNA (Pimentel et al. 2005). These
results suggested a functional coupling between the
replication and parD modules in plasmid R1.
The RNase activity of Kid toxin
The discovery that RelE toxin belonging to the relBE
system inhibits translation by cleaving the mRNA on
the ribosome (Pedersen et al. 2003), triggered a
search for similar activities in the toxins of other
TAS. It was found that Kid/PemK, the identical toxin
of the parD/pem systems of plasmid R1 and R100
(Tsuchimoto et al. 1988), are specific
endoribonucleases that, in contrast to RelE, can
cleave RNA in the absence of ribosomes (Zhang et al.
2003, Zhang et al. 2004, Munoz-Gomez et al. 2004,
Munoz-Gomez et al. 2005). This cleavage inhibits the
potential of the cells to synthesize proteins. Kid toxin
cleaves RNA as RNase A or RNase T1 do (Kamphuis
et al. 2006). This cleavage is initiated by a
nucleophilic attack of the scissile phosphate by the
oxygen of an adjacent 2'-OH residue and requires a
catalytic acid, a catalytic base and additional
stabilizing interactions. The structure of the functional
dimeric Kid toxin is available (Hargreaves et al.
2002) and the model of a structure of the complex of
Kid with a mimetic RNA substrate based in NMR
data has been proposed (Kamphuis et al. 2006) (see
Fig. 1). Residues proposed by this model to play key
roles in RNA cleavage and in specific interactions
with the substrate have been evaluated using a
collection of specific mutants in the toxin
(Diago-Navarro et al. 2009a). The results of these
studies have supported the predictions of the model.
Kis/PemI and MazE antitoxins are able to neutralize
the RNase activity of their respective toxins in
addition to their potential in inhibiting protein
synthesis (Munoz-Gomez et al. 2004, 2005, Zhang et
al. 2003, 2004). The available structures of the
MazF2-MazE2-MazF2 heterohexamer (Kamada et al.
2003) and of the Kid2-Kis2-Kid2 heterohexamer,
modelled on the MazE-MazF heterohexamer
(Kamphuis et al. 2007b), explain this neutralization as
the result of an interference of the C-terminal region
of the antitoxin with toxin residues involved in RNA
binding or cleavage. Neutralization of Kid RNase
activity by the Kis antitoxin occurs either in
prokaryotic or eukaryotic cells (de la Cueva-Mendez
et al. 2003, Munoz-Gomez et al. 2005).
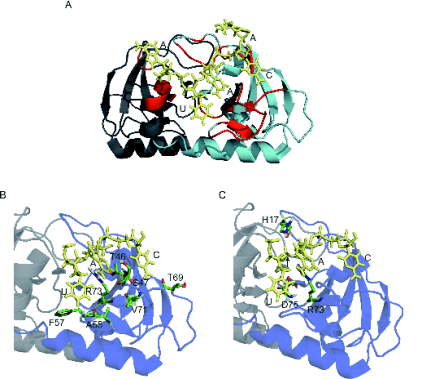
Fig. 1. Modelled mechanism of binding and cleavage of RNA by Kid toxin. A) RNA binding pocket of Kid. Kid monomers
are coloured in grey and cyan. Residues involved in one of the symmetric RNA binding surfaces are shown in red. B) and C)
show respectively Kid residues (sticks) involved in specific interactions with the RNA core sequence and these forming the active
site of the protein. The arrow points to the scissile phosphate. RNA bases (AUACA) are coloured in pale yellow.
The RNase activity of Kid toxin explains several
other results obtained before this primary activity was
discovered: the recovery of compromised plasmid R1
replication was due to the RNase activity of the Kid
toxin (Pimentel et al. 2005), and the inhibition of
ColE1 replication by the Kid toxin observed in vitro
and in vivo (Potrykus et al. 2002, Munoz-Gomez et al.
2005) could be explained by the cleavage of the RNA
transcript required to make the RNA that primes
replication of this plasmid. This cleavage was shown
using purified RNA ColE1 pre-primer as a substrate
of Kid; however a detailed analysis of this cleavage
occurring during transcription remains to be carried
out. The ability of Kid to inhibit cell growth/viability
both in prokaryotes and eukaryotes is related to the
inhibition of protein synthesis due to its RNase
activity (Munoz-Gomez et al. 2005).
Interestingly the Kid toxin shows structural
homology to the CcdB toxin (Hargreaves et al. 2002)
even if CcdB rather than cleaving RNA targets DNA
gyrase (Bernard and Couturier 1992).
Regulation of the parD system
TAS are subjected to strict control to prevent the
deleterious effects of their toxins. The activity of the
toxins is neutralized by direct interactions with their
antitoxins (see above). Interestingly these interactions
lead to complexes that regulate the operon and
maintain, the toxin-antitoxin transcripts at low levels.
Recent studies of the transcriptional regulation of
parD showed that dimers of the antitoxin pilot the
interactions of the repressor complex with specific
sequences of the promoter-operator region and that
the relative levels of Kis and Kid proteins influence
this regulation: a hetero-octameric complex formed in
excess of the antitoxin binds in the promoter-operator
region efficiently, suggesting that it is involved in
repression, and a heterohexameric Kis-Kid complex
formed in excess of the toxin is the main species
involved in toxin neutralization (Monti et al. 2007,
Kamphuis et al. 2007b). This complex binds with low
affinity to the promoter-operator region thus
favouring deregulation of the operon and reposition of
the antitoxin levels. The interplay of the two proteins
could restore the regulated situation when the
antitoxin decays. This hypothesis explains the
requirement of two different proteins to regulate the
system.
Post-transcriptional regulatory mechanisms
(Ruiz-Echevarria et al. 1995a) contribute to maintain,
under standard growth conditions, an A/T ratio close
to 2 that results in a repressed situation. The selective
activity of the Lon protease on the Kis antitoxin
(Tsuchimoto et al. 1992) can unbalance this situation.
RelBE TYPE SYSTEMS: PROTEIN
INHIBITION BY CLEAVAGE OF mRNA ON
THE RIBOSOMES
One of the most studied TAS members is the relBE
gene, present in the chromosome of many eu- and
archae- bacteria and firstly reported in E. coli
(Christensen et al. 2001). Homologues were found in
Bacteria, Archaea (Gerdes et al. 2005, Pandey and
Gerdes 2005), and in plasmids (Gronlund and Gerdes
1999, Hayes 2003). In addition to the E. coli operon
(Gotfredsen and Gerdes 1998), the nature as bona fide
TAS relBE loci has been confirmed for
S. pneumoniae (Nieto et al. 2006), Mycobacterium
tuberculosis (Korch et al. 2009), Pyrococcus
horikoshii (Takagi et al. 2005), and Methanococcus
jannaschii (Francuski and Saenger 2009). In E. coli
cultures subjected to nutritional stress, especially in
conditions that impair protein synthesis, transcription
of relBE increased and toxin RelE was activated due
to degradation of its cognate RelB antitoxin by the
Lon protease. This resulted in cell growth arrest,
concomitantly with inhibition of translation
(Christensen et al. 2001, Gotfredsen and Gerdes,
2002). Over-expression of EcRelE induced stasis
from which cells can recover by antitoxin production
(Pedersen et al. 2002). A similar behaviour was found
in E. coli by activation of the pneumococcal SpRelE
toxin but, in this case, prolonged exposure of the cells
to the toxin led to cell death rather than to cell stasis
(Nieto et al. 2006). Therefore, chromosomal relBE
could act as a stress response locus more than a cell
killing system, adjusting the rates of protein synthesis
under unfavourable growth conditions.
RelE mRNA cleavage mechanism
RelE was shown to inhibit translation by cleaving
mRNAs, both in vivo and in vitro by RelE positioning
at the ribosomal A site (Christensen and Gerdes
2003, Pedersen et al. 2003). Determination of the
RelE structure showed that the protein has a shape
similar to the C-terminal region of the translation
elongation factor EF-G (Takagi et al. 2005), thus
allowing its access into the A site of the ribosome.
RelE-mediated cleavage occurred preferentially
between the second and the third bases of stop and
sense codons with a G at third position (Pedersen et
al. 2003). In agreement with the entrance of RelE in
the ribosomal A-site was the finding that the release
factor I (RFI), which binds to the ribosomal A site in
the translation termination stage prevented in vitro
cleavage of mRNA mediated by RelE (Pedersen et al.
2003). RFI-mutants showed increased sensitivity to
the RelE and Kid toxins (Diago-Navarro et al.
2009b). In the case of RelE, overproduction of
tmRNA was able to rescue stalled ribosomes, thus
counteracting its toxic effect (Christensen and Gerdes
2003). EcRelE only cleaved ribosome-bound mRNAs
(Christensen and Gerdes 2003, Pedersen et al. 2003).
However, E. coli strains with deletions in genes
encoding toxins (RelE, MazF, ChpBK, YoeB, YafQ
and YhaV) were shown not to be involved in
cleavage of a particular mRNA composed of rare Arg
codons that caused ribosome pausing (Garza-Sanchez
et al. 2008); in addition, arginine starvation was
shown to induce mRNA cleavage at specific codons.
However, cleavage occurred with the same specificity
in the strain lacking the toxins, indicating that mRNA
cleavage occurring during arginine starvation was
independent of these known TA systems. Similar
results were found by Aiba's group (Li et al. 2008b).
Interestingly, comparison of the fitness of two
isogenic E. coli strains, one wild type (wt) and the
other having deletions in five TAS (mazEF, relBE,
chpBK, yefM-YoeB, dinJ-yafQ) subjected to
short-term stress conditions (amino acid starvation,
acidic stress, antibiotic treatment, and long term
stationary phase) showed no significant differences
among them (Tsilibaris et al. 2007), suggesting that
TAS could be involved only in long-term evolution
(Van Melderen and Saavedra De Bast 2009).
Even though RelE has a microbial RNase fold it
lacked both basic and acidic catalytic residues
suggesting that they should be provided by the
ribosome (Buts et al. 2005, Condon 2006). The
solution of the three-dimensional structures of
EcRelE, alone and bound to programmed Thermus
thermophilus 70S ribosomes in both pre- and
post-cleavage states solved this puzzle (Neubauer,
2009). In the crystal, RelE occupies the A site,
establishing direct contacts with the 16S rRNA, and
thus preventing access of translation factors and
tRNA to the ribosome. The overall structure of RelE
hardly changed upon binding to the ribosome, but its
interaction with the A site on the 30S subunit seemed
to reorganize the mRNA, promoting a 2'-OH
hydrolysis between codon position two and three (Fig. 2). Considering the structural and functional
data, a model for RelE-mediated cleavage of mRNA
was proposed (Neubauer et al. 2009). Even though
cleavage of the mRNA on the ribosome is carried out
by RelE and not by the ribosome itself, this cell
particle is essential for the RelE catalysis.
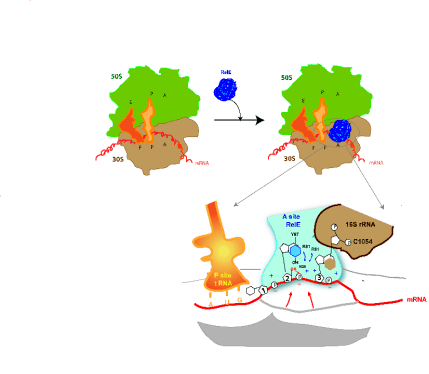
Fig. 2. Mechanism of cleavage of RNA on the ribosome. A) Schematic representation of RelE binding on the ribosome. E, P,
and A denotes specific ribosomal sites involved in translation. B) Model of endonucleolytic reaction of RelE on translated
mRNA. RelE approaches the mRNA to its active site through interactions between RelE basic side chains (+) (R45, R81, R61,
K52, K54, and R56) and the phosphate group in the mRNA (-). The stacking of the second nucleotide with tyrosine 87(Y87)
and of the third nucleotide with the residue C1054 of 16S rRNA are indicated. The red arrow indicates the nucleophilic attack
of 2'O- on the phosphate group. The water molecule and residues R61 and R81 present in the active sites of RelE are also shown.
In addition to the E. coli RelE, the structure of the
complex RelE-RelB, has been determined for
P. horikoshii (PhRelBE) (Takagi et al. 2005) and
M. jannaschii (MjRelBE) (Francuski and Saenger
2009). PhRelBE forms a tetrameric complex, in
which a molecule of RelB wraps around a compact
RelE dimer generating a heterodimer. Two
heterodimers generate the heterotetramer through
interactions between PhRelB from one heterodimer
and PhRelE from another. The extensive wrapping of
RelB around RelE makes the dimer bigger, thus
hampering the entrance of RelE into the ribosome
A-site. MjRelBE forms also a heterotetrameric
complex with a different pattern. Data on the RelBE
complex from E. coli indicate that the C-terminal
region of RelB could be responsible for RelB-RelE
interaction. This region showed high sensitivity to
proteases, so that its interaction with RelE could
confer resistance to degradation; thus the RelB
C-terminal region could be involved in both
self-association and RelE binding (Cherny et al.
2007). NMR studies showed that RelB-RelE complex
provokes a helix displacement near the RelE mRNA
interferase active site resulting in the neutralization of
the positively charge catalytic sites by acidic residues
from RelB antitoxin (Li et al. 2009). The E. coli RelB
N-terminal domain may form a dimeric
ribbon-helix-helix structure (Cherny et al. 2007, Li et
al. 2008a), making it likely that the antitoxin would
use this motif for DNA binding (Li et al. 2008a,
Overgaard et al. 2008). However, the three-
dimensional structure of PhRelB showed the antitoxin
would recognize its DNA target as a dimer via a
leucine zipper motif (Takagi et al. 2005). In
conclusion, the structural data collected regarding
RelB indicate that despite the sequence and structure
homology between the toxins from E. coli, M.
jannaschii and P. horikoshii, the antitoxins may differ
in the mechanism they use to optimize their DNA
binding and their stability properties, adapting them
to their own host.
Auto-regulation
Two models have been proposed to explain the
mechanism of autorepression of the E. coli relBE
operon. The first considers the possibility that RelBE
complex could bind as a tetramer to the operator
sequence and that RelB has two levels of regulation
(Li et al. 2008a). The second model (Overgaard et al.
2008) proposes that RelBE complex could control
relBE transcription by binding to the DNA operator
as a trimer, RelB2-RelE. Thus, RelE would have two
RelB binding sites, one of low and another of high
affinity each playing different roles depending on the
toxin:antitoxin ratios (Overgaard et al. 2008).
YoeB mRNA interferase
Another mRNA interferase homologous to RelE,
YoeB, has been studied, albeit in less detail than
RelE. It has been grouped as belonging to the RelE
superfamily (Pandey and Gerdes 2005, Makarova et
al. 2009), and studied in E. coli and in other
microorganisms. Gene yoeB encodes toxin YoeB
belonging to the TA pair YefMYoeB, which was
identified as a homolog of the Txe toxin (Grady and
Hayes 2003) in E. coli. The Axe-Txe TA pair was
identified in a plasmid from Enterococcus faecium
(Grady and Hayes 2003), and homologs have been
studied in S. pneumoniae (Nieto et al. 2007),
Staphylococcus aureus (Yoshizumi et al. 2009), and
M. tuberculosis (Kumar et al. 2008). The structure of
the E. coli YoeB and the complex with its cognate
antitoxin, YefM, has been elucidated and the residues
involved in YoeB catalysis have been identified.
YoeB showed a structure similar to RNase Sa and
Barnase, two proteins with a characteristic microbial
RNase fold (Kamada and Hanaoka 2005); it also
exhibited similarities with RelE from E. coli and with
PhRelE monomer (Takagi et al. 2005, Neubauer et al.
2009). YoeB acts as a ribosome-dependent mRNA
interferase (Christensen-Dalsgaard and Gerdes 2008).
All experimental data suggest that YoeB inhibits
protein synthesis, but its primary function as a
blockade for this process is unclear. Whereas YoeB
might act as a sequence-specific endoribonuclease or
an mRNA interferase (Kamada and Hanaoka 2005,
Christensen-Dalsgaard and Gerdes 2008), it could be
that the endoribonuclease activity of YoeB is not
primarily required for its inhibitory function for
protein synthesis but to prevent the formation of the
initiation complex (Zhang and Inouye 2009). In this
latter case, YoeB would bind to the 50S ribosomal
subunit in 70S ribosomes, and would interact with the
A site, impairing formation of the initiation complex
and, as a consequence, inhibiting protein synthesis. In
turn, this inhibition would activate the latent
endoribonuclease activity of either ribosomes or
YoeB, resulting in cleavage of mRNA at the A site.
Interaction of YoeB with its cognate antitoxin YefM
could induce a conformational rearrangement of the
RNase catalytic site of YoeB leading to the
movement of some of the residues involved in
catalysis, away from the active site. This
conformational change in the catalytic site of YoeB
could explain the mechanism employed by YefM to
neutralize YoeB toxicity. Regulation of the
expression of this TAS is similar to the other
examples mentioned: YefM is the primary repressor
and YoeB acts as a repressor enhancer (Kedzierska et
al. 2007). However, this picture might be an
oversimplification of the regulatory circuit (W. T.
Chang and C. C. Yeo, personal communication).
SOME POSSIBLE BIOMEDICAL APPLICATIONS/IMPLICATIONS OF TAS
The structural and functional information available on
TA interactions in several systems make it possible to
search for or to design molecules able to interfere
with these interactions and trigger the activity of the
toxin. BRET (Bioluminiscence Resonance Energy
Transfer) technology has been used to monitor
toxin-antitoxin interactions (Nieto et al. 2006, Lioy et
al. 2010). These assays could act as a powerful tool in
the search for possible new antibiotics against cells
containing endogenous or acquired toxin-antitoxin
systems. The role of some TAS, particularly, the
HipBA system of E. coli in bacterial persistence has
been pointed out (Moyed and Bertrand 1983). A
10,000 increase in the numbers of persistent cells was
associated with a mutation, hipA7, in the toxin gene.
However, in spite of the well characterized
mechanism of action of the toxin and of its
neutralization by the antitoxin (Schumacher et al.
2009), the pathway involved in the activation of this
epigenetic phenomenon, affecting a small fraction of
the population, is not well understood. Persistent cells
have important implications in combating antibiotic
resistances in the clinical context as these cells are
tolerant to antibiotics (Bigger 1944).
TAS were first characterized as virulence-
associated determinants on the basis of their
prevalence in the chromosomes of virulent versus
avirulent strains of specific human pathogens (Hopper
et al. 2000, Daines et al. 2007). Moreover, TAS are
widely conserved in plasmids from common hospital
pathogens, including vancomycin-resistant
enterococci (VRE) and aminoglycoside-resistant
E. coli (Moritz and Hergenrother 2007, Perichon et al.
2008). The sequenced strains of Mycobacterium
tuberculosis have 60 TAS including 7 homologues of
mazEF (Pandey and Gerdes 2005). The contribution
of these systems to the pathogenic character of this
micro-organism remains to be fully explored. It is
noteworthy that in bacterial species such as Vibrio
cholerae, up to 13 TAS cluster in a mega-integron
structure (Pandey and Gerdes 2005). Thus TAS could
contribute to the genetic stability of mobile genetic
elements and could play a role in stabilizing virulent
traits during the evolution of pathogenic bacteria.
Measurable attenuation of the virulence of pathogenic
bacteria through the genetic inactivation of TAS using
suitable animal infection models remains to be
provided to properly address the possible role of TAS
during infection.
Even before RNA was identified as the direct
target of Kid, the bacterial Kid toxin was known to
prevent proliferation in eukaryotic cells including
yeast, oocites of Xenopus laevis and HeLa cells (de la
Cueva-Mendez et al. 2003). Kid induced apoptosis in
the later cells and the antitoxin Kis was protecting
from these effects. This indicates that regulated
expression of Kid and Kis might be used to kill
particular cells, including tumour cell lines, in a
selective way. This may be achieved by expressing
the kid and kis genes under the control of promoters
that are, respectively, induced and repressed in these
cells, and that have the inverse behaviour in normal
cells. Since Kid also inhibits the growth of embryonic
cells, a similar strategy might be used to prevent the
growth of particular cell lineages during development.
This approach could have value in studies of
differentiation, organogenesis or degenerative
disorders (de la Cueva-Mendez et al. 2003). Indeed
differential expression of the toxin and antitoxin
genes of the parD system in fertilized embryos of
zebrafish has been used to eliminate selectively the
germ line of this fish and to study its role in sex
differentiation (Slanchev et al. 2005). Similarly the
RelE protein from E. coli acts as a global inhibitor of
translation not only in bacteria or archaea but also in
eukaryotic cells. In Saccharomyces cerevisiae the
expression of the bacterial relE gene is toxic to yeast
cells (Kristoffersen et al. 2000), and in a human
osteosarcoma cell line the toxin RelE retards growth
and leads to cell death by apoptosis (Yamamoto et al.
2002).
The growing structural and functional information
on toxin-antitoxin systems open important avenues to
the exploration of their biomedical and
biotechnological implications and applications both in
prokaryotic and eukaryotic cells.
ACKNOWLEDGEMENTS
The authors acknowledge the financial support of the
MICINN, the Spanish Ministry of Science and
Innovation, (CSD2008-00013; BFU2008-
00179-E/BMC) and the dedication of many
colleagues and friends that contributed to these
studies over the years.
REFERENCES
Bernard P, Couturier M: Cell killing by the F plasmid CcdB protein involves poisoning of DNA-topoisomerase II complexes. J Mol Biol 226:735-745, 1992.
Bigger J: Treatment of Staphylococcal infections with penicillin by intermittent sterilisation. Lancet 244:497-500, 1944.
Bravo A, de Torrontegui G, Diaz R: Identification of components of a new stability system of plasmid R1, ParD, that is close to the origin of replication of this plasmid. Mol Gen Genet 210:101-110, 1987.
Bravo A, Ortega S, de Torrontegui G, Diaz R: Killing of Escherichia coli cells modulated by components of the stability system ParD of plasmid R1. Mol Gen Genet 215:146-151, 1988.
Buts L, Lah J, Dao-Thi MH, Wyns L, Loris R: Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem Sci 30:672-679, 2005.
Cherny I, Overgaard M, Borch J, Bram Y, Gerdes K, Gazit E: Structural and thermodynamic characterization of the Escherichia coli RelBE toxin-antitoxin system: indication for a functional role of differential stability. Biochemistry 46:12152-12163, 2007.
Christensen SK, Gerdes K: RelE toxins from Bacteria and Archaea cleave mRNAs on translating ribosomes, which are rescued by tmRNA. Mol Microbiol 48:1389-1400, 2003.
Christensen SK, Mikkelsen M, Pedersen K, Gerdes K: RelE, a global inhibitor of translation, is activated during nutritional stress. Proc Natl Acad Sci USA 98:14328-14333, 2001.
Christensen-Dalsgaard M, Gerdes K: Translation affects YoeB and MazF messenger RNA interferase activities by different mechanisms. Nucleic Acids Res 36:6472-6481, 2008.
Condon C: Shutdown decay of mRNA. Mol Microbiol 61:573-583, 2006.
Daines DA, Wu MH, Yuan SY: VapC-1 of nontypeable Haemophilus influenzae is a ribonuclease. J Bacteriol 189: 5041-5048, 2007.
de la Cueva-Mendez G, Mills AD, Clay-Farrace L, Diaz-Orejas R, Laskey RA: Regulatable killing of eukaryotic cells by the prokaryotic proteins Kid and Kis. EMBO J 22:246-251, 2003.
Diago-Navarro E, Kamphuis MB, Boelens R, Barendregt A, Heck AJ, van den Heuvel RH, Diaz-Orejas R: A mutagenic analysis of the RNase mechanism of the bacterial Kid toxin by mass spectrometry. The FEBS J 276:4973-4986 2009a.
Diago-Navarro E, Mora L, Buckingham RH, Diaz-Orejas R, Lemonnier M: Novel Escherichia coli RF1 mutants with decreased translation termination activity and increased sensitivity to the cytotoxic effect of the bacterial toxins Kid and RelE. Mol Microbiol 71:66-78, 2009b.
Francuski D, Saenger W: Crystal structure of the antitoxin-toxin protein complex RelB-RelE from Methanococcus jannaschii. J Mol Biol 393: 898-908, 2009.
Garza-Sanchez F, Gin JG, Hayes CS: Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol 378:505-519, 2008.
Gerdes K, Rasmussen PB, Molin S: Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci USA 83:3116-3120, 1986.
Gerdes K, Christensen SK, Lobner-Olesen A: Prokaryotic toxin-antitoxin stress response loci. Nature Rev Microbiol 3:371-382, 2005.
Gotfredsen M, Gerdes K: The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol 29:1065-1076, 1998.
Gotfredsen M, Gerdes K: Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J Bacteriol 182:561-572, 2002.
Grady R, Hayes F: Axe-Txe, a broad-spectrum proteic toxin-antitoxin system specified by a multidrug-resistant, clinical isolate of Enterococcus faecium. Mol Microbiol 47:1419-1432, 2003.
Gronlund H, Gerdes K: Toxin-antitoxin systems homologous with relBE of Escherichia coli plasmid P307 are ubiquitous in prokaryotes. J Mol Biol 285:1401-1415, 1999.
Hargreaves D, Santos-Sierra S, Giraldo R, Sabariegos-Jareno R, de la Cueva-Mendez G, Boelens R, Diaz-Orejas R, Rafferty JB: Structural and functional analysis of the kid toxin protein from E. coli plasmid R1. Structure 10:1425-1433, 2002.
Hayes F: Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499, 2003.
Hiraga S, Jaffe A, Ogura T, Mori H, Takahashi H: F plasmid ccd mechanism in Escherichia coli. J Bacteriol 166:100-104, 1986.
Hopper S, Wilbur JS, Vasquez BL, Larson J, Clary S, Mehr IJ, Seifert HS, So M: Isolation of Neisseria gonorrhoeae mutants that show enhanced trafficking across polarized T84 epithelial monolayers. Infection Immun 68:896-905, 2000.
Kamada K, Hanaoka F: Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol Cell 19:497-509, 2005.
Kamada K, Hanaoka F, Burley SK: Crystal structure of the MazE/MazF complex: molecular bases of antidote-toxin recognition. Mol Cell 11:875-884, 2003.
Kamphuis MB, Bonvin AM, Monti MC, Lemonnier M, Munoz-Gomez A, van den Heuvel RH, Diaz-Orejas R, Boelens R: Model for RNA binding and the catalytic site of the RNase Kid of the bacterial parD toxin-antitoxin system. J Mol Biol 357:115-126, 2006.
Kamphuis MB, Monti MC, van den Heuvel RH, Lopez-Villarejo J, Diaz-Orejas R, Boelens R: Structure and function of bacterial kid-kis and related toxin-antitoxin systems. Protein Pept Lett 14:113-124, 2007a.
Kamphuis MB, Monti MC, van den Heuvel RH, Santos-Sierra S, Folkers GE, Lemonnier M, Diaz-Orejas R, Heck AJ, Boelens R: Interactions between the toxin kid of the bacterial parD system and the antitoxins Kis and MazE. Proteins 67:219-231, 2007b.
Kedzierska B, Lian LY, Hayes F: Toxin-antitoxin regulation: bimodal interaction of YefM-YoeB with paired DNA palindromes exerts transcriptional autorepression. Nucleic Acids Res 35:325-339, 2007.
Korch SB, Contreras H, Clark-Curtiss JE: Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J Bacteriol 191:1618-1630, 2009.
Kristoffersen P, Jensen GB, Gerdes K, Piskur J: Bacterial toxin-antitoxin gene system as containment control in yeast cells. Appl Environ Microbiol 66:5524-5526, 2000.
Kumar P, Issac B, Dodson EJ, Turkenburg JP, Mande SC: Crystal structure of Mycobacterium tuberculosis YefM antitoxin reveals that it is not an intrinsically unstructured protein. J Mol Biol 383:482-493, 2008.
Li GY, Zhang Y, Inouye M, Ikura M: Structural mechanism of transcriptional autorepression of the Escherichia coli RelB/RelE antitoxin/toxin module. J Mol Biol 380:107-119, 2008a.
Li GY, Zhang Y, Inouye M, Ikura M: Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J Biol Chem 284:14628-14636, 2009.
Li X, Yagi M, Morita T, Aiba H: Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol Microbiol 68:462-473, 2008b.
Lioy VS, Rey O, Balsa D, Pellicer T, Alonso JC: A toxin-antitoxin module as a target for antimicrobial development. Plasmid 63:31-39, 2010.
Makarova KS, Wolf YI, Koonin EV: Comprehensive comparative-genomic analysis of Type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol Direct 4:19, 2009.
Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E: chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol 175:6850-6856, 1993.
Monti MC, Hernandez-Arriaga AM, Kamphuis MB, Lopez-Villarejo J, Heck AJ, Boelens R, Diaz-Orejas R, van den Heuvel RH: Interactions of Kid-Kis toxin-antitoxin complexes with the parD operator-promoter region of plasmid R1 are piloted by the Kis antitoxin and tuned by the stoichiometry of Kid-Kis oligomers. Nucleic Acids Res 35:1737-1749, 2007.
Moritz EM, Hergenrother PJ: Toxin-antitoxin systems are ubiquitous and plasmid-encoded in vancomycin-resistant enterococci. Proc Natl Acad Sci USA 104:311-316, 2007.
Moyed HS, Bertrand KP: hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J Bacteriol 155:768-775, 1983.
Munoz-Gomez AJ, Santos-Sierra S, Berzal-Herranz A, Lemonnier M, Diaz-Orejas R: Insights into the specificity of RNA cleavage by the Escherichia coli MazF toxin. FEBS Lett 567:316-320, 2004.
Munoz-Gomez AJ, Lemonnier M, Santos-Sierra S, Berzal-Herranz A, Diaz-Orejas R: RNase/anti-RNase activities of the bacterial parD toxin-antitoxin system. J Bacteriol 187:3151-3157, 2005.
Neubauer C, Gao YG, Andersen KR, Dunham CM, Kelley AC, Hentschel J, Gerdes, Ramakrishnan V, Brodersen DE: The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell 139:1084-1095, 2009.
Nieto C, Pellicer T, Balsa D, Christensen SK, Gerdes K, Espinosa M: The chromosomal relBE2 toxin-antitoxin locus of Streptococcus pneumoniae: characterization and use of a bioluminescence resonance energy transfer assay to detect toxin-antitoxin interaction. Mol Microbiol 59:1280-1296, 2006.
Nieto C, Cherny I, Khoo SK, de Lacoba MG, Chan WT, Yeo CC, Gazit E, Espinosa M: The yefM-yoeB toxin-antitoxin systems of Escherichia coli and Streptococcus pneumoniae: functional and structural correlation. J Bacteriol 189:1266-1278, 2007.
Overgaard M, Borch J, Jorgensen MG, Gerdes K: Messenger RNA interferase RelE controls relBE transcription by conditional cooperativity. Mol Microbiol 69:841-857, 2008.
Pandey D, Gerdes K: Toxin- antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res 55:78-89, 2005.
Pedersen K, Christensen SK, Gerdes K: Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol Microbiol 45:501-510, 2002.
Pedersen K, Zavialov AV, Pavlov MY, Elf J, Gerdes K, Ehrenberg M: The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140, 2003.
Perichon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M: Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob Agents Chemother 52:2581-2592, 2008.
Pimentel B, Madine MA, de la Cueva-Mendez G: Kid cleaves specific mRNAs at UUACU sites to rescue the copy number of plasmid R1. EMBO J 24:3459-3469, 2005.
Potrykus K, Santos S, Lemonnier M, Diaz-Orejas R, Wegrzyn G: Differential effects of Kid toxin on two modes of replication of lambdoid plasmids suggest that this toxin acts before, but not after, the assembly of the replication complex. Microbiology 148:2489-2495, 2002.
Ruiz-Echevarria MJ, de la Cueva G, Diaz-Orejas: Translational coupling and limited degradation of a polycistronic messenger modulate differential gene expression in the parD stability system of plasmid R1. Mol Gen Genet 248:599-609, 1995a.
Ruiz-Echevarria MJ, de la Torre MA, Diaz-Orejas R: A mutation that decreases the efficiency of plasmid R1 replication leads to the activation of parD, a killer stability system of the plasmid. FEMS Microbiol Lett 130:129-135, 1995b.
Santos-Sierra S, Giraldo R, Diaz-Orejas R: Functional interactions between homologous conditional killer systems of plasmid and chromosomal origin. FEMS Microbiol Lett 152:51-56, 1997.
Santos-Sierra S, Giraldo R, Diaz-Orejas R: Functional interactions between chpB and parD, two homologous conditional killer systems found in the Escherichia coli chromosome and in plasmid R1. FEMS Microbiol Lett 168:51-58, 1998.
Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG: Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396-401, 2009.
Slanchev K, Stebler J, de la Cueva-Mendez G, Raz E: Development without germ cells: the role of the germ line in zebrafish sex differentiation. Proc Natl Acad Sci USA 102:4074-4079, 2005.
Takagi H, Kakuta Y, Okada T, Yao M, Tanaka I, Kimura M: Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat Struct Mol Biol 12:327-331, 2005.
Tsilibaris V, Maenhaut-Michel G, Mine N, Van Melderen L: What is the benefit to Escherichia coli of having multiple toxin-antitoxin systems in its genome? J Bacteriol 189:6101-6108, 2007.
Tsuchimoto S, Ohtsubo H, Ohtsubo E: Two genes, pemK and pemI, responsible for stable maintenance of resistance plasmid R100. J Bacteriol 170:1461-1466, 1988.
Tsuchimoto S, Nishimura Y, Ohtsubo E: The stable maintenance system pem of plasmid R100: degradation of PemI protein may allow PemK protein to inhibit cell growth. J Bacteriol 174:4205-4211, 1992.
Van Melderen L, Saavedra De Bast M: Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet 5:e1000437, 2009.
Yamamoto TA, Gerdes K, Tunnacliffe A: Bacterial toxin RelE induces apoptosis in human cells. FEBS Lett 519:191-194, 2002.
Yoshizumi S, Zhang Y, Yamaguchi Y, Chen L, Kreiswirth BN, Inouye M: Staphylococcus aureus YoeB homologues inhibit translation initiation. J Bacteriol 191:5868-5872, 2009.
Zhang J, Zhang Y, Zhu L, Suzuki M, Inouye M: Interference of mRNA function by sequence-specific endoribonuclease PemK. J Biol Chem 279:20678-20684, 2004.
Zhang Y, Inouye M: The inhibitory mechanism of protein synthesis by YoeB, an Escherichia coli toxin. J Biol Chem 284:6627-6638, 2009.
Zhang Y, Zhang J, Hoeflich KP, Ikura M, Qing G, Inouye M: MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli. Mol Cell 12:913-923, 2003.
|
BACK
|