Journal of APPLIED BIOMEDICINE
ISSN 1214-0287 (on-line)
ISSN 1214-021X (printed)
Volume 8 (2010), No 4, p 189-198
DOI 10.2478/v10136-009-0026-4
Temperature-perception, molecules and mechanisms
Rafael Catala, Julio Salinas
Address: Julio Salinas, Department of Environmental Biology, Centro de Investigaciones Biologicas (CIB - CSIC)
Ramiro de Maeztu 9, 28040 Madrid, Spain
salinas@cib.csic.es
Received 5th March 2010.
Revised 11th May 2010.
Published online 24th May 2010.
Full text article (pdf)
Abstract in xml format
Summary
Key words
Introduction
Physiological changes induced by low temperature
The process of cold acclimation
Adaptive responses to low temperature in other organisms
Conclusions and perspectives
Acknowledgements
References
SUMMARY
The strategies used by living organisms to survive under low and freezing temperatures reveal the extraordinary adaptability of life on Earth. Understanding the molecular mechanisms underlying cold adaptation and freezing survival will provide new insights into the existing relationships between living organisms and their environment, and the possibility of developing multiple biotechnological applications. In the case of plants, the use of classical genetic and new "omics" approaches is allowing to the identification of new elements involved in regulating the cold acclimation response. The challenge ahead is to determine temperature-perception molecules and mechanisms, to uncover new internodes of multiple responses, and to integrate the regulation not only at the transcriptome but also at proteome and metabolome levels. Attaining these goals will significantly contribute global understanding the adaptive strategies plants have evolved to cope with hostile environmental conditions, and to the development biotechnological strategies to improve crop tolerance to freezing and other important abiotic stresses.
KEY WORDS
Arabidopsis; low temperature response; cold acclimation; freezing tolerance; cold signalling
INTRODUCTION
Low and freezing temperatures severely conditioned
life of all organisms in many different ways, from the
reduction of biochemical reaction rates or changes in
membrane fluidity and protein conformation, to the
need for protection against freezing. Regarding
plants, freezing temperatures limit their development
and geographical distribution, and account for
important losses in crop production. In temperate
regions, many species have evolved a low
temperature response, known as cold acclimation,
whereby they can increase their freezing tolerance
after a period of exposure to low-nonfreezing
temperatures (Levitt 1980). Understanding the
molecular mechanisms governing this adaptive
response is important in learning how plants grow and
reproduce under hostile conditions, and should help
to develop biotechnological strategies to improve
crop tolerance to freezing. The process of cold
acclimation is quite complex and involves a high
number of physiological and biochemical changes,
most of them controlled by low temperatures through
changes in gene expression, which during cold
acclimation is mainly regulated at the transcriptional
level (Salinas 2002, Yamaguchi-Shinozaki and
Shinozaki 2006, Van Buskirk and Thomashow 2006),
although recent findings indicate that regulation at the
posttranscriptional, translational and posttranslational
levels may also be important (Mazzucotelli et al.
2006, Zhu et al. 2007, Catala and Salinas 2008). For
some years, our lab has been using the model plant
Arabidopsis and a collection of experimental
approaches to uncover signalling intermediates
involved in regulating plant responses to low
temperature. Some of them resulted in the mediation
of metabolic adjustments or changes in the
composition of plasma membrane induced by cold
conditions. Furthermore, by screening a cDNA library
from cold-acclimated seedlings of Arabidopsis with
a subtracted probe enriched in cold-induced
transcripts (Jarillo et al. 1994), we identified a series
of Rare Cold Inducible (RCI) genes that have been
shown to be implicated in cold signalling. The
functional characterization of these genes has
provided important insights into the molecular
mechanisms underlying plant responses to low
temperature, including cold acclimation. A number of
reviews covering plant responses to low temperature
and cold acclimation have been recently published
(Mazzucotelli et al. 2006, Van Buskirk and
Thomashow 2006, Yamaguchi-Shinozaki and
Shinozaki 2006, Zhu et al. 2007, Catalá and Salinas
2008, Penfield 2008). In this paper, we will describe
and discuss some of our contributions to the
understanding of how plants respond when facing low
temperature conditions, and how the process of cold
acclimation is regulated.
PHYSIOLOGICAL CHANGES INDUCED BY LOW TEMPERATURE
One of the primary effects of low temperature in cells
is the alteration of membrane fluidity properties and
fatty acid composition due to an increase in the
content of polyunsaturated lipids (Murata and Los
1997, Aguilar et al. 1998, Suzuki et al. 2001,
Hayward et al. 2007). In plants, this increase is
essential for low-temperature survival and directly
affects membrane-bound metabolic processes such as
respiration and photosynthesis (Cossins 1994). A shift
toward an anaerobic metabolism has been reported in
different species (i.e., maize and rice) when exposed
to low temperature, which provokes a rapid increase
of alcohol dehydrogenase gene (ADH) expression and
activity (Christie et al. 1991). In Arabidopsis, we
have demonstrated that ADH expression is also
induced by cold stress, and that this induction is
mediated by abscisic acid (ABA) (Jarillo et al. 1993).
Nevertheless, in spite of the induction, our results
revealed that ADH is not essential for the correct
development of cold acclimation response (Jarillo et
al. 1993).
Photoinhibition, a reduction of photosynthetic
activity that may be caused by a combination of light
and cold, has been directly correlated with low
temperature sensitivity due to an increase in
photoxidative stress (Harvaux and Kloppstech 2001).
Anthocyanins, which function in photoprotection
acting as light-screening pigments, accumulate in
leaves and stems in response to low temperature and
changes in light intensity (Mancinelli 1984, Christie
et al. 1994). The synthesis of these pigments is
controlled, in part, by the key enzymes phenyl
ammonia lyase (PAL) and chalcone synthase (CHS).
In our laboratory, Leyva et al. (1995) reported that the
expression of Arabidopsis PAL and CHS genes is
induced in response to cold in a light-dependent
manner, and that this induction is regulated at the
transcriptional level. As in the case of ADH, genetic
analysis has revealed that CHS is not essential for the
increase in freezing tolerance originated by cold
acclimation (Leyva et al. 1995). Taken together, all
these results suggest that the function of ADH, PAL
and CHS would be related more to the response to
chilling stress than to cold acclimation. Low
temperature also affects the protein composition of
photosystems (Gray et al. 1997, Krol et al. 1999). We
could show that the expression of CAB1, a gene
encoding a light-harvesting chlorophyll a/b binding
protein, is induced when Arabidopsis plants are
exposed to 4 °C (Capel et al. 1998). In contrast to
PAL and CHS, the induction of CAB1 in response to
cold is light-independent (Capel et al. 1998),
indicating that it is regulated through a different
pathway. Interestingly, it has been reported that
Arabidopsis light-harvesting complex (LHC) proteins,
including CAB1, play an important role in protecting
photosystem I from photoinhibition (Alboresi et al.
2009).
In addition of modifying the content of
polyunsaturated lipids in the plasma membrane, low
temperature also induces changes in the composition
of plasma membrane proteins. Kawamura and
Uemura (2003) reported the isolation of 38 proteins
from Arabidopsis plasma membrane whose levels
increase or decrease in response to cold. Among
them, they identified proteins involved in CO2
fixation, membrane repair, protection of membrane to
osmotic stress, or proteolysis (Kawamura and Uemura
2003). One of these proteins, with function in
membrane repairing, SYT1, has been demonstrated to
play a role in freezing tolerance and cold acclimation
(Schapire et al. 2008, Yamazaki et al. 2008).
Recently, we described a family of eight Arabidopsis
small hydrophobic proteins (AtRCI2A-H) that are
also located in the plasma membrane (Medina et al.
2007). The expression of AtRCI2 genes is
differentially regulated in Arabidopsis organs and in
response to low temperature (Capel et al. 1997,
Medina et al. 2001, 2007). AtRCI2A-C and AtRCI2H
are able to complement for the loss of the yeast
AtRCI2A-homologue PMP3 (Medina et al. 2007),
which has been involved in the maintenance of
plasma membrane potential, suggesting a similar role
for AtRCI2 during cold stress (Medina et al. 2007).
It has been well known for several years that
exposition to low temperatures also provokes changes
in the composition of cell wall components in several
plant species. Wei et al. (2006) reported that
expression of C3H, a Rhododendron gene encoding
a protein implicated in lignin accumulation in the cell
wall, is cold induced, suggesting a possible role of
lignin in low temperature responses. On the same
way, we reported the identification of RCI3 gene of
Arabidopsis, that encodes an active cationic
peroxidase, a kind of protein that has been involved
in lignin and suberin depositions in the cell wall
(Llorente et al. 2002).
THE PROCESS OF COLD ACCLIMATION
As already mentioned, cold acclimation is an adaptive
response whereby some plants acquire an increase in
freezing tolerance upon a prior low-nonfreezing
temperature exposition (Levitt 1980). Plants are able
to acclimate to cold by extensively reprogramming
their transcriptome, proteome and metabolome. In
recent years, the efforts of several laboratories,
including our own, have began to elucidate the
regulatory networks that mediate cold acclimation. In
the next paragraphs, we intend to present the progress
in our understanding of how low temperature signals
are perceived and transduced leading to the activation
of this adaptive response.
Signal transduction in cold acclimation
The first step in any signalling pathway implies the
recognition of the signal by a receptor. It has been
proposed that changes in the fluidity of cell
membranes could act as a receptor of temperature
changes in plant cells (Orvar et al. 2000, Sangwan et
al. 2002). Nevertheless, experimental evidence
demonstrating that this is the case has not yet been
provided, and how plants perceive low temperatures
still remains to be elucidated. What seems to be clear
is that transient increases in cytosolic calcium
concentration ([Ca2+]cyt) are essential for plant
response to low temperature (Knight 2000). Increases
in [Ca2+]cyt mainly result from Ca2+ influx through
permeable channels in the plasma membrane and/or
Ca2+ discharge from internal stores (Piñeros and
Tester 1997). After Ca2+ influx, efflux systems to
internal stores and out of the cell restore [Ca2+]cyt to
unstimulated levels via Ca2+ pumps and Ca2+/H+
exchangers (Knight 2000). One of the RCI genes
identified in our lab, RCI4, turned out to be identical
to CAX1, an Arabidopsis gene encoding a H+/Ca2+
antiporter essential for correct regulation of [Ca2+]cyt
(Hirschi et al. 1996). RCI4/CAX1 expression is
induced by cold through an ABA-independent
pathway (Catala et al. 2003). Functional analysis
revealed that RCI4/CAX1 is involved in the increase
of freezing tolerance that is induced by cold
acclimation (Catala et al. 2003). In fact, rci4/cax1
mutants show enhanced capacity to cold acclimate,
which correlates with a higher induction of
CBF/DREB1 genes and their corresponding targets in
response to low temperature (see below) (Catalá et al.
2003). We proposed that RCI4/CAX1 would function
to restore [Ca2+]cyt to unstimulated levels after the
increase induced by low temperatures (Fig. 1). The
absence of RCI4/CAX1 activity would result in an
extended cold signal that would provoke a higher
induction of the CBFs/DREB1s and their target genes,
and, therefore, an increase in Arabidopsis freezing
tolerance. These results demonstrate that an accurate
[Ca2+]cyt regulation is essential for the correct
development of cold acclimation response, and that
RCI4/CAX1 is an essential partner of the regulatory
mechanisms controlling [Ca2+]cyt homeostasis in
Arabidopsis.
Generally, Ca2+ signals mediating regulatory
pathways are associated with reversible
phosphorylation events. Reversible phosphorylation
of pre-existing proteins has been demonstrated to be
essential for cold acclimation in alfalfa and
Arabidopsis (Tahtiharju et al. 1997, Monroy et al.
1998). RCI1A and RCI1B are two Rare Cold Inducible
genes of Arabidopsis identified in our laboratory that
encode two members of the 14-3-3 protein family
(Jarillo et al. 1994, Abarca et al. 1999). The 14-3-3
proteins function as dimmers to regulate the activity
of other previously phosphorylated proteins to which
they interact (Abarca et al. 1999, Roberts et al. 2002).
The kinetics of RCI1A and RCI1B expression in
response to low temperature correlates with the
increase of freezing tolerance induced by cold
acclimation, suggesting a role for these proteins in
this adaptive process (Jarillo et al. 1994). The 14-3-3
protein family of Arabidopsis includes 12 isoforms,
but only RCI1A and RCI1B are cold inducible (Jarillo
et al. 1994, Roberts et al. 2002). Preliminary data with
a null mutant for RCI1A indicate that the
corresponding protein acts as a negative regulator of
cold acclimation and is involved in regulating
cold-induced gene expression (Catala et al.
unpublished).
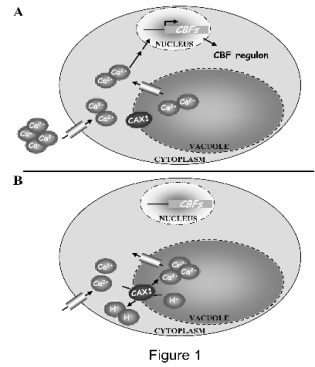
Fig. 1. Functional model of RCI4/CAX1 during cold acclimation. A. Exposition to low temperature induces a transient
increase in [Ca2+]cyt as a consequence of Ca2+ influx through permeable channels in the plasma membrane and discharge from internal stores. This increase, conveniently decoded and transduced by downstream effectors, enhances the expression of the CBF
genes and their targets, activating the cold acclimation response. B. When the cold signal vanishes, RCI4/CAX1 introduces
cytoplasmic Ca2+ into the vacuole, contributing, together with other Ca2+ transporters, to restore [Ca2+]cyt to unstimulated levels.
The restoration of [Ca2+]cyt levels restrains induction of CBFs.
Regulation of gene expression during cold acclimation
Global expression analyses have revealed that about
one thousand genes are regulated by low temperature
in Arabidopsis (Lee et al. 2005, Matsui et al. 2008,
Zeller et al. 2009). Among these genes, one third is
repressed, with only one gene encoding a
transcription factor, which indicates that gene
expression during cold acclimation is mainly
activated (Lee et al. 2005). In addition, many early
cold-induced genes encode transcription factors or
proteins involved in transcription. Thus, more than
one hundred genes have been annotated to function in
transcription, 95 of them coding for transcription
factors (Lee et al. 2005). These results indicate that
cold-induced gene expression is regulated through
different signal transduction pathways. To date, the
most important pathways that have been identified
and characterized are those mediated by ABA and the
CBF/DREB1 transcription factors, respectively.
ABA and cold-induced gene expression
It is well documented that ABA levels increase in
plants in response to different environmental adverse
conditions, including low temperatures, and that
exogenous treatment with ABA enhances freezing
tolerance (Chen and Gusta 1983, Lang et al. 1989).
Moreover, it has been described that ABA-defective
or -insensitive Arabidopsis mutants have a lower
capacity to cold acclimate, indicating that ABA is
essential for a full cold acclimation response (Heino
et al. 1990, Gilmour and Thomashow 1991). In our
laboratory, Llorente et al. (2000) isolated a mutant of
Arabidopsis, named freezing sensitive 1 (frs1), that
showed reduced freezing tolerance before and after
cold acclimation. frs1 resulted to be an allele of
ABA3, which encodes an important component of
ABA biosynthesis. Actually, frs1 mutants have lower
ABA levels compared to wild-type plants. The fact
that frs1 is affected in its constitutive freezing
tolerance indicates that ABA not only plays an
important role in cold acclimation but also has a
function in the constitutive tolerance of Arabidopsis
to freezing temperatures (Llorente et al. 2000).
Molecular characterization of frs1 demonstrated that
ABA mediates the induction of gene expression in
response to low temperatures (Llorente et al. 2000).
Confirming our results, Xiong et al. (2001) reported
the identification and characterization of los5, which
resulted to be another allele of ABA3. Similar to frs1
mutants, los5 plants have reduced levels of ABA,
decreased accumulation of transcripts corresponding
to different cold-inducible genes, and reduced
tolerance to freezing (Xiong et al. 2001). All these
data demonstrate that ABA mediates cold acclimation
by controlling cold-regulated gene expression.
The CBF family of transcriptional activators
The best characterized signalling pathway controlling
cold-inducible gene expression in Arabidopsis is that
mediated by the small family of transcriptional
activators named C-repeat binding
factors/dehydration responsive element binding
factors 1 (CBFs/DREB1s). These factors regulate the
expression of around 12% of the Arabidopsis
cold-inducible genes (Fowler and Thomashow 2002),
which gives an idea about their significance in cold
acclimation. CBF1 was the first to be isolated due to
its ability to bind to the COR15A promoter and to
induce its expression (Stockinger et al. 1997).
Subsequently, three different labs, including our own,
independently reported that CBF1 belongs to a
protein family composed by three members (CBF1-3)
(Gilmour et al. 1998, Liu et al. 1998, Medina et al.
1999). CBFs have an acidic C-terminal region that
acts as a transcriptional activator motif and an AP2
DNA-biding domain to recognize and interact with
the C-repeat (CRT)/dehydration responsive element
(DRE) motif (CCGAC) present in the promoters of a
number of cold-inducible genes that constitute the
CBF regulon (Stockinger et al. 1997, Gilmour et al.
1998, Liu et al. 1998). The three CBF genes are
organized in tandem in chromosome 4 of Arabidopsis
and their expression is rapidly and transiently induced
by cold (Gilmour et al. 1998, Liu et al. 1998, Medina
et al. 1999). Using a quantitative trait loci (QTL)
mapping approach, we determined that the genetic
basis of the natural variation for freezing tolerance in
Arabidopsis is mainly produced by a QTL that
colocalize with the CBF cluster (Alonso-Blanco et al.
2005), confirming the relevance of these factors in
cold acclimation. Constitutive overexpression of each
individual CBF gene in Arabidopsis induces the
accumulation of mRNAs from genes belonging to the
CBF regulon as well as an increase in freezing
tolerance, suggesting that the CBFs might be
functionally redundant (Kasuga et al. 1999, Gilmour
et al 2004). Genetic analysis, however, uncovered a
different and more interesting scenario. In fact, the
molecular and functional characterization of a
knock-out mutant for CBF2 revealed that the absence
of CBF2 increases Arabidopsis tolerance to freezing
temperatures, before and after cold acclimation
(Novillo et al. 2004). Furthermore, this increase
correlates with a higher accumulation of CBF1 and
CBF3 transcripts, and, consequently, of mRNAs
corresponding to CBF-target genes, under both
control and low-temperature conditions (Novillo et al.
2004). These data indicate that CBF2 negatively
modulates the expression of CBF1 and CBF3,
ensuring the adequate expression of the CBF-regulon
during cold acclimation (Novillo et al. 2004). On the
other hand, the characterization of Arabidopsis plants
with reduced induction of CBF1 or CBF3 in response
to low temperature revealed that, contrary to CBF2,
CBF1 and CBF3 are not involved in regulating the
expression of other CBF genes, demonstrating that
the three CBFs do not have the same function (Fig. 2)
(Novillo et al. 2007). Moreover, results showed that
although CBF1 and CBF3 seem to positively regulate
the induction of the same target genes, they are
concertedly required to induce the whole
CBF-regulon and, therefore, the complete
development of the cold acclimation response in
Arabidopsis (Novillo et al. 2007).
The results described above evidence the high
complexity to which the regulation of CBF expression
is subjected, and prompt the importance of an
accurate control of their induction levels. Different
transcription factors have been reported as interacting
with the promoters of the CBF genes and regulating
their induction (Fig. 2). Thus, MYB15 binds to the
CBF promoters to repress their expression (Agarwal
et al. 2006). The ICE1 MYC-like bHLH transcription
factor binds to the CBF3 promoter to activate its
expression (Chinnusamy et al. 2003). Moreover, ICE1
interacts to and represses MYB15 (Agarwal et al.
2006). Vogel et al. (2005) reported that the
overexpression of an Arabidopsis zinc finger, ZAT12,
significantly dumped the induction of the CBFs in
response to low temperature. Strikingly, ZAT12 does
not seem to affect CBF-targeted gene expression,
which suggests a complex role for this protein in the
regulation of cold-inducible gene expression and cold
acclimation (Vogel et al. 2005). ICE2, another
MYC-like bHLH transcription factor that activates
CBF1 expression (Fursova et al. 2009), has also been
described. In addition to these factors, other proteins
involved in regulating the expression of CBFs have
been identified (Zhu et al. 2007), confirming that
these genes are, in fact, subjected to a tight regulation.
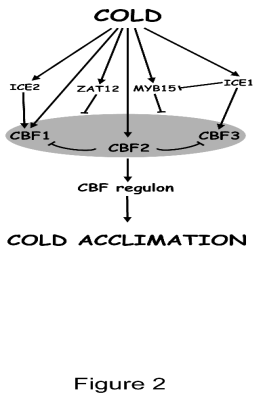
Fig. 2. CBFs are central factors in cold-acclimation.
CBFs are subjected to a tightly regulation. ICE1 and ICE2
have been described to positively regulate CBF3 and CBF1
expression, respectively. ZAT12 and MYB15 negatively
regulate CBF expression. In addition, CBF2 represses
CBF1 and CBF3 expression. ICE1 has also been described
to repress MYB15. The expression of CBFs leads to the
induction of the CBF regulon and the subsequent activation
of cold acclimation response.
Interplay between cold acclimation and plant
responses to other abiotic stresses
It has already been mentioned that about one
thousand genes are regulated by low temperature in
Arabidopsis (Lee et al. 2005, Matsui et al. 2008,
Zeller et al. 2009). Interestingly, many of these genes
are also regulated by salt and water stresses (Matsui
et al. 2008, Zeller et al. 2009), indicating that plant
responses to abiotic stresses are closely related and
share common components. Ishitani et al. (1997),
trying to dissect the complex network that regulates
Arabidopsis response to cold and osmotic stresses,
screened for Arabidopsis mutants with deregulated
expression of a luciferase reporter gene driven by the
promoter of RD29A, a CBF-target gene whose
expression is induced in response to cold,
dehydration, high salt and ABA. This genetic
approach allowed them to isolate and characterize a
number of mutants (hos and los) with altered capacity
to cold acclimate. Most of these mutants were also
affected in their tolerance to water and/or salt stress
(Xiong et al. 2002), supporting the idea that plant
responses to low temperature, water stress and high
salt have several features in common. Using a similar
experimental approach, we identified Arabidopsis
mutants with high (hor) or low (lor) expression of
AtRCI2A (see above) in response to low temperature,
dehydration, high salt or ABA (Medina et al. 2005).
As in the case of hos and los mutants, several hor and
lor mutant lines are not only affected in their ability
to cold acclimate but also in their sensitivity to
dehydration, high salt and ABA treatments (Medina
et al. 2005), further supporting the narrow
relationship that exists between plant responses to
abiotic stresses. Based on the results obtained from
the physiological and molecular characterization of
the hor and lor mutants, we proposed a working
model that reflects the remarkable complexity of the
networks whereby plants appropriately respond when
exposed to adverse abiotic environmental conditions
(Fig. 3).
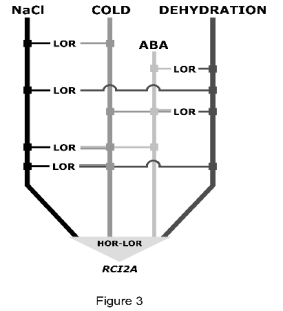
Fig. 3. Working model for interactions between plant
responses to cold, dehydration, high salt and ABA
treatments. From the results obtained when characterizing
the hor and lor mutants, a signalling network involved in
abiotic stress response can be proposed. The pathways
mediating AtRCI2A expression in response to cold,
dehydration, salt stress and ABA treatment interact and
converge at different levels. Indeed, we found lor mutants
affected in the expression of RCI2A in response to two
(Cold/NaCl; Cold/Dehydration; NaCl/Dehydration;
ABA/Dehydration), three (Cold/NaCl/Dehydration;
Cold/Dehydration/ABA; Cold/NaCl/ABA) and all
treatments. We also identified hor mutants altered in RCI2A
expression by all treatments. The proteins defined by the
mutants identified are placed in putative positions in the
pathways. Nevertheless, the existence of individual
pathways mediating single responses, as well as the
possibility that this signalling network interacts with other
networks cannot be excluded.
ADAPTIVE RESPONSES TO LOW TEMPERATURE IN OTHER ORGANISMS
Many microorganisms and animals have also evolved
adaptive mechanisms to cope with low and freezing
temperatures, being able to successfully colonize cold
environments, which represent most of the Earth
biosphere. Interestingly, a number of these adaptive
mechanisms are common with plants. For instance,
animals and microorganisms also preserve membrane
function and fluidity under low temperature
conditions by increasing fatty acid unsaturation
(Aguilar et al. 1998, Suzuki et al. 2001, Hayward et
al. 2007). Moreover, as in the case of plants, animals
and microorganisms produce high levels of
compatible solutes, i.e. sugars, amino acids, polyols,
to protect cells against the deleterious effects
originated by freezing tempertures (Yancey 2005).
Intracellular ice formation is lethal and most
organisms living in regions exposed to subzero
temperatures, including plants, synthesize antifreeze
proteins to inhibit, slow down, or control the growth
of ice crystals (Venketesh and Dayananda 2008).
Antioxidant defense is another common adaptive
mechanism evolved by microorganisms, plants and
animals to survive at low temperature. Strategies for
the detoxification of cold-induced reactive oxygen
species that cause membrane damage include the
production of high levels of antioxidant enzymes and
detoxifying compounds (Schulze et al. 2005,
Voituron et al. 2005, Storey 2006, Gocheva et al.
2009).
CONCLUSIONS AND PERSPECTIVES
Low temperatures are one of the most important plant
abiotic stress factors. Confronted with them, some
plants are able to adapt through mechanisms based on
membrane composition changes, protein synthesis
and activation of active oxygen scavenging systems.
One of the most interesting adaptive responses to low
temperature which plants have evolved is the so
called process of cold acclimation, whereby they
acquire an increase in freezing tolerance upon a prior
low-nonfreezing temperature treatment. This adaptive
mechanism mainly relies on gene induction, although
recent reports indicate postranscriptional, translational
and posttranslational regulation is also involved and
therefore identification of intermediaries in protein
and mRNA transport and stability will be essential in
order to completely understand the regulation of the
cold acclimation process. How plants perceive the
cold signal and how the signal is transduced to
accurately activate plant responses, including the
process of cold acclimation, still remains largely
unknown. Recent studies in our own and others'
laboratories have identified signalling components
that mediate cold acclimation. Interestingly, some of
these works have reported that several regulators are
shared with other abiotic environmental responses.
These discoveries reveal the complexity of molecular
mechanisms underlying the adaptation of plants to
their environment, raising the question of how plants
coordinate the signalling pathways involved in the
response to different abiotic stresses.
ACKNOWLEDGEMENTS
Work in our laboratory is supported by grants
GEN2006-27787-E/VEG, BIO2007-65284 and
CSD2007-00057 from the Spanish Ministry of
Science and Innovation, and grant P2006/GEN-0191
from the Regional Goverment of Madrid.
REFERENCES
Abarca D, Madueno F, Martinez-Zapater JM, Salinas J: Dimerization of Arabidopsis 14-3-3 proteins: structural requirements within the N-terminal domain and effect of calcium. FEBS Lett 462:377-382, 1999.
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK: A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636-37645, 2006.
Aguilar PS, Cronan JE, Jr., de Mendoza D: A Bacillus subtilis gene induced by cold shock encodes a membrane phospholipids desaturase. J Bacteriol 180:2194-2200, 1998.
Alboresi A, Ballottari M, Hienerwadel R, Giacometti GM, Morosinotto T: Antenna complexes protect Photosystem I from photoinhibition. BMC Plant Biol 9:article 71, 2009.
Alonso-Blanco C, Gomez-Mena C, Llorente F, Koornneef M, Salinas J, Martinez-Zapater JM: Genetic and molecular analyses of natural variation indicate CBF2 as a candidate gene for underlying a freezing tolerance quantitative trait locus in Arabidopsis. Plant Physiol 139:1304-1312, 2005.
Capel J, Jarillo JA, Salinas J, Martinez-Zapater JM: Two homologous low-temperature-inducible genes from Arabidopsis encode highly hydrophobic proteins. Plant Physiol 115:569-576, 1997.
Capel J, Jarillo JA, Madueno F, Jorquera MJ, Martinez-Zapater JM, Salinas J: Low temperature regulates Arabidopsis Lhcb gene expression in a light-independent manner. Plant J 13:411-418, 1998.
Catala R, Salinas J: Regulatory mechanisms involved in cold acclimation response. Span J Agric Res 6:211-220, 2008.
Catala R, Santos E, Alonso JM, Ecker JR, Martinez-Zapater JM, Salinas J: Mutations in the Ca2+/H+ transporter CAX1 increase CBF/DREB1 expression and the cold-acclimation response in Arabidopsis. Plant Cell 15:2940-2951, 2003.
Chen TH, Gusta LV: Abscisic acid-Induced freezing resistance in cultured plant cells. Plant Physiol 73:71-75, 1983.
Chinnusamy V, Ohta M, Kanrar S, Lee BH, Hong X, Agarwal M, Zhu JK: ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev 17:1043-1054, 2003.
Christie PJ, Hahn M, Walbot V: Low-temperature accumulation of alcohol dehydrogenase-1 mRNA and protein activity in maize and rice seedlings. Plant Physiol 95:699-706, 1991.
Christie PJ, Alfenito MR, Walbot V: Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 194:541-549, 1994.
Cossins AR: Temperature Adaptation in Biological Membranes. Portland Press, London 1994, pp. 63-76.
Fowler S, Thomashow MF: Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675-1690, 2002.
Fursova OV, Pogorelko GV, Tarasov VA: Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 15:98-103, 2009.
Gilmour SJ, Thomashow MF: Cold acclimation and cold-regulated gene expression in ABA mutants of Arabidopsis thaliana. Plant Mol Biol 17:1233-1240, 1991.
Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF: Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J 16:433-442, 1998.
Gilmour SJ, Fowler SG, Thomashow MF: Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767-781, 2004.
Gocheva YG, Tosi S, Krumova ET, Slokoska LS, Miteva JG, Vassilev SV, Angelova MB: Temperature downshift induces antioxidant response in fungi isolated from Antarctica. Extremophiles 13:273-281, 2009.
Gray GR, Chauvin LP, Sarhan F, Huner N: Cold acclimation and freezing tolerance (A complex interaction of light and temperature). Plant Physiol 114:467-474, 1997.
Harvaux M, Kloppstech K: The protective functions of carotenoid and flavonoid pigments against excess visible radiation at chilling temperature investigated in Arabidopsis npq and tt mutants. Planta 213:953-966, 2001.
Hayward SAL, Murray PA, Gracey AY, Cossins AR: Beyond the lipid hypothesis: Mechanisms underlying plasticity in inducible cold tolerance. Adv Exp Med Biol 594:132-142, 2007.
Heino P, Sandman G, Lang V, Nordin K, Palva ET: Abscisic acid deficiency prevents development of freezing tolerance in Arabidopsis thaliana (L.) Heynh. Theor Appl Genet 79:801-806, 1990.
Hirschi KD, Zhen RG, Cunningham KW, Rea PA, Fink GR: CAX1, an H+/Ca2+ antiporter from Arabidopsis. Proc Natl Acad Sci USA 93:8782-8786, 1996.
Ishitani M, Xiong L, Stevenson B, Zhu JK: Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: interactions and convergence of abscisic acid-dependent and abscisic acid-independent pathways. Plant Cell 9:1935-1949, 1997.
Jarillo JA, Leyva A, Salinas J, Martinez-Zapater JM: Low temperature induces the accumulation of alcohol dehydrogenase mRNA in Arabidopsis thaliana, a chilling-tolerant plant. Plant Physiol 101:833-837, 1993.
Jarillo JA, Capel J, Leyva A, Martinez-Zapater JM, Salinas J: Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol Biol 25:693-704, 1994.
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K: Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287-291, 1999.
Kawamura Y, Uemura M: Mass spectrometric approach for identifying putative plasma membrane proteins of Arabidopsis leaves associated with cold acclimation. Plant J 36:141-154, 2003.
Knight H: Calcium signalling during abiotic stress in plants. Int Rev Cytol 195:269-324, 2000.
Krol M, Ivanov AG, Jansson S, Kloppstech K, Huner NP: Greening under high light or cold temperature affects the level of xanthophyll-cycle pigments, early light-inducible proteins, and light-harvesting polypeptides in wild-type barley and the chlorina f2 mutant. Plant Physiol 120:193-204, 1999.
Lang V, Heino P, Palva ET: Low temperature acclimation and treatment with exogenous abscisic acid induce common polypeptides in Arabidopsis thaliana (L.) Heinh. Theor Appl Genet 77:729-734, 1989.
Lee BH, Henderson DA, Zhu JK: The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. Plant Cell 17:3155-3175, 2005.
Levitt J: Responses of Plants to Environmental Stresses: chilling, freezing and high temperatures stresses. Academic Press, New York, 1980.
Leyva A, Jarillo JA, Salinas J, Martinez-Zapater JM: Low temperature Induces the Accumulation of Phenylalanine Ammonia-Lyase and Chalcone Synthase mRNAs of Arabidopsis thaliana in a Light-Dependent Manner. Plant Physiol 108:39-46, 1995.
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K: Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391-1406, 1998.
Llorente F, Oliveros JC, Martinez-Zapater JM, Salinas J: A freezing-sensitive mutant of Arabidopsis, frs1, is a new aba3 allele. Planta 211:648-655, 2000.
Llorente F, Lopez-Cobollo RM, Catala R, Martinez-Zapater JM, Salinas J: A novel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J 32:13-24, 2002.
Mancinelli AL: Photoregulation of anthocyanin synthesis: VIII. Effect of light pretreatments. Plant Physiol 75:447-453, 1984.
Matsui A, Ishida J, Morosawa T, Mochizuki Y, Kaminuma E, Endo TA, Okamoto M, Nambara E, Nakajima M, Kawashima M, Satou M, Kim JM, Kobayashi N, Toyoda T, Shinozaki K, Seki M: Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol 49:1135-1149, 2008.
Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A: The E3 ubiquitin ligase gene family in plants: regulation by degradation. Current Genomics 7:509-522, 2006.
Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J: The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiol 119:463-470, 1999.
Medina J, Catala R, Salinas J: Developmental and stress regulation of RCI2A and RCI2B, two cold-inducible genes of Arabidopsis encoding highly conserved hydrophobic proteins. Plant Physiol 125:1655-1666, 2001.
Medina J, Rodriguez-Franco M, Penalosa A, Carrascosa MJ, Neuhaus G, Salinas J: Arabidopsis mutants deregulated in RCI2A expression reveal new signalling pathways in abiotic stress responses. Plant J 42:586-597, 2005.
Medina J, Ballesteros ML, Salinas J: Phylogenetic and functional analysis of Arabidopsis RCI2 genes. J Exp Bot 58:4333-4346, 2007.
Monroy AF, Sangwan V, Dhindsa RS: Low temperature signal transduction during cold acclimation: protein phophatase 2A as an early target for cold-inactivation. Plant J 13:653-660, 1998.
Murata N, Los DA: Membrane fluidity and temperature perception. Plant Physiol 115:875-879, 1997.
Novillo F, Alonso JM, Ecker JR, Salinas J: CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DREB1A expression and plays a central role in stress tolerance in Arabidopsis. Proc Natl Acad Sci USA 101:3985-3990, 2004.
Novillo F, Medina J, Salinas J: Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc Natl Acad Sci USA 104:21002-21007, 2007.
Orvar BL, Sangwan V, Omann F, Dhindsa RS: Early steps in cold sensing by plant cells: the role of actin cytoskeleton and membrane fluidity. Plant J 23:785-794, 2000.
Penfield S: Temperature perception and signal transduction in plants. New Phytol 179:615-628, 2008.
Pineros M, Tester M: Characterization of the high-affinity verapamil binding site in a plant plasma membrane Ca2+-selective channel. J Membr Biol 157:139-145, 1997.
Roberts MR, Salinas J, Collinge DB: 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol Biol 50:1031-1039, 2002.
Salinas J: Molecular mechanisms of signal transduction in cold acclimation. In Hames BD, Glover DM: Frontiers in Molecular Biology, Oxford University Press, Oxford 2002, pp: 116-139.
Sangwan V, Orvar BL, Beyerly J, Hirt H, Dhindsa RS: Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J 31:629-638, 2002.
Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, Salinas J, Mancuso S, Valpuesta V, Baluska F, Botella MA: Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20:3374-3388, 2008.
Schulze ED, Beck E, Muller-Hohenstein K: Plant Ecology. Springer, Berlin/Heidelberg, 2005.
Stockinger EJ, Gilmour SJ, Thomashow MF: Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035-1040, 1997.
Storey KB: Reptile freeze tolerance: metabolism and gene expression. Cryobiology 52:1-16, 2006.
Suzuki I, Kanesaki Y, Mikami K, Kanehisa M, Murata N: Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol Microbiol 40:235-244, 2001.
Tahtiharju S, Sangwan V, Monroy AF, Dhindsa RS, Borg M: The induction of kin genes in cold-acclimating Arabidopsis thaliana. Evidence of a role for calcium. Planta 203:442-447, 1997.
Van Buskirk HA, Thomashow MF: Arabidopsis transcription factors regulating cold acclimation. Physiol Plant 126:72-80, 2006.
Venketesh S, Dayananda C: Properties, potentials and prospects of antifreezing proteins. Crit Rev on Biotechnol 28:57-82, 2008.
Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF: Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195-211, 2005.
Voituron Y, Servais S, Romestaing C, Douki T, Barre H: Oxidative DNA damage and antioxidant defenses in the European common lizard (Lacerta vivipara) in supercooled and frozen states. Cryobiology 51:74-82, 2005.
Wei H, Dhanaraj AL, Arora R, Rowland LJ, Fu Y, Sun L: Identification of cold acclimation-responsive Rhododendron genes for lipid metabolism, membrane transport and lignin biosynthesis: importance of moderately abundant ESTs in genomic studies. Plant Cell Environ 29:558-570, 2006.
Xiong L, Ishitani M, Lee H, Zhu JK: The Arabidopsis LOS5/ABA3 locus encodes a molybdenum cofactor sulfurase and modulates cold stress- and osmotic stress-responsive gene expression. Plant Cell 13:2063-2083, 2001.
Xiong L, Schumaker KS, Zhu JK: Cell signalling during cold, drought, and salt stress. Plant Cell 14 (Suppl):S165-183, 2002.
Yamaguchi-Shinozaki K, Shinozaki K: Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781-803, 2006.
Yamazaki T, Kawamura Y, Minami A, Uemura M: Calcium-dependent freezing tolerance in Arabidopsis involves membrane resealing via synaptotagmin SYT1. Plant Cell 20:3389-3404, 2008.
Yancey PH: Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208:2819-2830, 2005.
Zeller G, Henz SR, Widmer CK, Sachsenberg T, Ratsch G, Weigel D, Laubinger S: Stress-induced changes in the Arabidopsis thaliana transcriptome analyzed using whole-genome tiling arrays. Plant J 58:1068-1082, 2009.
Zhu J, Dong CH, Zhu JK: Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10:290-295, 2007.
|
BACK
|