Journal of APPLIED BIOMEDICINE
ISSN 1214-0287 (on-line)
ISSN 1214-021X (printed)
Volume 9 (2011), No 3, p 163-171
DOI 10.2478/v10136-009-0037-1
Docking studies and effects of syn-anti isomery of oximes derived from pyridine imidazol bicycled systems as potential human
acetylcholinesterase reactivators
Ana Paula Guimaraes, Tanos Celmar Costa Franca, Teodorico Castro Ramalho, Magdalena Nascimento Renno, Elaine Fontes Ferreira da Cunha, Karina
Silvia Matos, Daiana Teixeira Mancini, Kamil Kuca
Address: Teodorico Castro Ramalho, Chemistry Department - Federal University of Lavras - Campus Universitario, 3037, 37200-000, Lavras, MG, Brazil
teo@dqi.ufla.br
Received 15th November 2010.
Revised 10th February 2011.
Published online 7th April 2011.
Full text article (pdf)
Abstract in xml format
Summary
Key words
Introduction
Materials and methods
Results and discussion
Conclusion
Acknowledgments
References
SUMMARY
In order to contribute to a better understanding of the mechanism of action of oximes, we evaluated the affinities of 10 new oximes, derived from
pyridine-imidazol bicycled systems, for human acetylcholinesterase (HssAChE) complexed with tabun, by estimating their docking energy
values and comparing of the values obtained to known oximes using the software Molegro Virtual Docker (MVD)®. We evaluated the influence of the
position of the oxime group as substituent in the structures and, also, the influence of the oxime group syn-anti isomery on the docking
score values for all the molecules studied. Results suggest that: the affinities of the 10 new oximes for the tabun inhibited HssAChE
active site are better than pralidoxime’s and similar to trimedoxime’s; the meta-pralidoxime could have more affinity for the
HssAChE active site and the oximes’ anti isomers could present slightly better affinities for the HssAChE active site than
the syn isomers.
KEY WORDS
acetylcholinesterase; docking studies; oximes; neurotoxic agents; theoretical calculation
INTRODUCTION
Despite the existence of many different oximes in use
today against intoxication with neurotoxic agents, the
literature has not yet reported one able to act
effectively against all the existing neurotoxic agents
and oximes usually effective with one specific nerve
agent can be completely ineffective with another.
This probably happens because their mechanisms of
action are not yet well elucidated (Ekstrom et al.
2007). In addition, other relevant factors: the
adequate orientation of the phosphoryl bond inside
the active site, the most suitable oxime charge, the
most adequate angles for attacking the phosphylated
serine, the influence of the oxime's isomery, and the
chemical environment of the oxime group, remain
unknown despite the fact that they are recurrent issues
in the literature (Bay et al. 1958, Smirnova et al.
1975, Castro and Figueroa-Villar 2002, Bartosova et
al. 2005, Goncalves et al. 2006, 2010, Ekstrom et al.
2007, Worek et al. 2007, Delfino and Figueroa-Villar
2009, Delfino et al. 2009, Ramalho et al. 2010).
In the present work, in order to contribute to a
better understanding of the mechanism of action of
oximes (Fig. 1), we proposed the structures and
evaluated in silico the affinities of 10 new oximes,
derived from pyridine-imidazol bicycled systems, for
the HssAChE active site inhibited by the neurotoxic
agent tabun. The softwares MVD® and Spartan® were
used to estimate the values of the oximes' affinity
(measured by the docking scores). The 10 oximes
were studied together with the standard oximes
pralidoxime (2-PAM), trimedoxime (TMB-4), and
obidoxime as references. Additionally the reactivation
constants of 2PAM and its ortho, meta and para
isomers were calculated according to a procedure
formerly established by Ramalho et. al. (2010). We
evaluated the influence of the oxime group position as
a substituent in the structures for 2-PAM and the
10 new oximes and, also, the influence of the oximes
syn-anti isomery in the docking scores values for all
the molecules studied. Our results suggest that the
affinities of the 10 new oximes for the HssAChE
active site are better than all 2PAM's isomers and
some were quite similar to TMB-4's. It was also
observed that meta-2-PAM could present better
affinities for the HssAChE active site than para and
ortho-2-PAM, and that 62.5% of the anti isomers
presented slightly better affinities for the HssAChE
active site when compared to the syn isomers.
MATERIALS AND METHODS
Docking energy calculations
The structure of the HssAChE used was that
phosphonylated by tabun proposed and optimized by
Goncalves et al. (2006) and complexed with
toxogonine, using as template the structure reported
by Kryger et al. (2000) deposited in the Proteins Data
Bank (Bernstein et al. 1977, Berman et al. 2000)
under the PDB code IB41. The water molecules were
removed using the program MVD®, the
three-dimensional structures of the oximes (Fig. 2)
were built and optimized with the software Spartan
Pro 5® and their partial atomic charges calculated by
the PM3 semi-empirical method. The compounds
were docked into the HssAChE binding site using
MVD® (Thomsen et al. 2006) according to
instructions which considered all the protein residues
as flexible. Binding sites were restricted within
spheres with radius of 8 Å, centered at the toxogonine
binding site in the protein complex and enclosing all
the active site residues. Due to the stochastic nature of
the ligand-protein docking search algorithm, about 10
runs were conducted and 30 docking solutions were
retained for each ligand. The best superimposing
poses related to toxogonine, were chosen to the
analysis performed in this work.
DFT Studies
QM/MM techniques were performed to determine the
preferred route for the reactivation process. On the
technical side, we applied a procedure combining
docking technique and DFT calculations at the
QM/MM interface for the enzymatic mechanism. The
QM calculations were carried out in the Spartan08
(Hehre et al. 1999) and Gaussian98 (Frisch et al.
2001) packages. The QM region, which consisted of
residues, neighbouring peptide bonds, link atoms,
crystallographic water molecules, ligand and
inhibitor, was confined into a sphere with a radius of
15 Å, centered at each oxime.
The initial coordinates for the heavy atoms were
taken from the HssAChE 3D structure proposed by
Goncalves et al. (2006). All the transition states,
intermediates and precursors involved were
calculated. Each conformer was fully optimized at the
DFT level with B3LYP/6-31G (Ramalho and Taft
2005). Furthermore, after each optimization, a force
constant calculation was made in order to verify
whether the optimized structures were indeed local
minima (no imaginary frequencies) or transition states
(one imaginary frequency).
RESULTS AND DISCUSSION
Docking results
The cavity (Fig. 3) of the HssAChE active site was
calculated by MVD® as having 949,696 Å3. The
results of the docking studies of the oximes studied
inside this cavity, allowed us to identify the relevant
H-Bonds that occur between each oxime and the
amino acid residues of the active site in order to
obtain the conformations adopted with these
molecules, compare them to the conformations of
toxogonine and thus get subsidies for the
investigations performed in this work.
The values of the docking energies obtained for
the best poses of the syn-anti isomers of the 10 new
oximes, the ortho, meta and para isomers of 2-PAM,
TMB-4 and obidoxime are presented in Fig. 4. Table
1 reports the H-bond energy values obtained for each
ligand in the HssAChE active site and, also, the
amino acid residues involved in H-bonds with them.
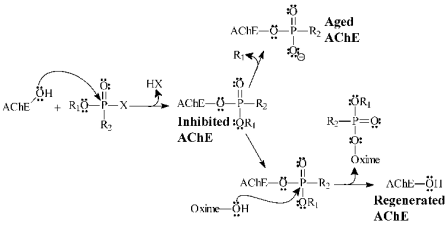
Fig. 1. Inhibition, desinhibition and ageing of acetylcholinesterase. X is the leaving group.
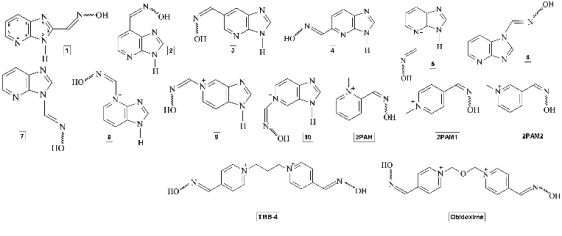
Fig. 2. Structures of the oximes studied.
Fig. 3. Cavity of the active site of HssAChE complexed
with obidoxime.
An analysis of Fig. 4 indicates that all the 10 new
oximes isomers presented docking energies better
than the 2PAM isomers but worse than TMB-4 and
obidoxime isomers. This result reflects the smaller
size of 2PAM and the new oximes, avoiding a full
superposition to obidixime and interactions with the
totality of residues in the cavity (Figs 5 and 6).
However, oximes 2, 3, 5, 6 and 8 presented at least
one isomer with similar docking values to TMB-4
isomers.
H-bond energy values obtained for most of the
new oximes isomers are better than for 2PAM and
quite similar to the values obtained for TMB-4 and
obidoxime isomers (see Table 1). All the aminoacid
residues observed forming H-bonds with 2PAM were
also observed for the new oximes. The new oximes
also were able to form H-bonds with some additional residues not observed for 2PAM (Tyr68, Asp70,
Tyr120, Glu281 and Phe291). All the residues
observed making H-bonds with the new oximes were
observed in TMB-4 and obidoxime isomers which
were, also, able to make additional H-bonds with the
residues: Gly117, Gly118, Glu198, Ser199, Ala200
and His443 (see Table 1).
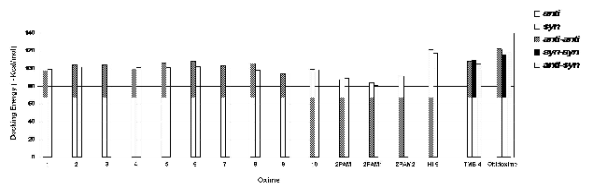
Fig. 4. Comparative docking energies of the syn-anti isomers of the new oximes, 2PAM isomers, HI-6, TMB-4 and
obidoxime.
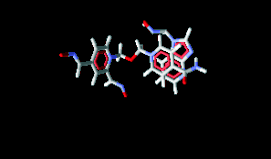
Fig. 5. Superposition of oxime 6 to obidoxime.
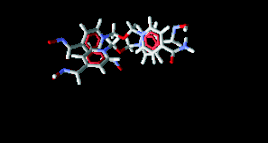
Fig. 6. Superposition of TMB-4 to obidoxime.
Concerning the position of the oxime group in
2PAM, the anti isomer of meta-2PAM presented a
slightly better docking energy value than para and
ortho-2PAM isomers but similar H-bond value to
ortho-2PAM. However the best result among the
2PAM syn isomers was observed for ortho-2PAM.
The presence of these amino acids with 2PAM,
2PAM1 and 2PAM2 compounds in anti and syn
conformations, suggests that they have a direct
influence on the mechanism of reactivation of
HssAChE inhibited by the organophosphorus agent
Tabun.
Finally, analysis of the effect of the syn-anti
isomery on the oximes' affinities for the HssAChE
active site showed that for 10 of the 16 oximes
studied (62.5%), the anti isomers presented better
docking values than the syn isomers. For the H-bonds
energy values the result pointed to 50%.
Reaction
The kinetics of the AChE reactivation process is
believed to occur in two steps: (1) the association of
the oxime to the inhibited AChE and (2) the
reactivation of AChE by the leaving of the oxime
complexed to the neurotoxic agent (Fig. 1). The
process of kinetic reactivation of AChE could be
illustrated by Equation 1:
K Rk R
EI + Ox EIOx E + I-Ox Eq. 1
Where EI is the organophosphate-inhibited
enzyme, Ox is the reactivator (oxime), E is the reactivated enzyme, EIOx is the intermediate complex
and I-Ox is the product. KR are the dissociation
constants, which represent the affinity of oximes for
tabun-inhibited AChE, and the rate constant for the
decomposition of the stable enzyme-inhibitor-reactivator complex, respectively.
Table 1. H-bond energy values in kcal/mol obtained and main aminoacid residues interacting with the ligands.
| Oxime |
Anti |
Syn |
Oxime |
Anti |
Syn | | 1 |
-7.2209
Phe291 (1)
Arg292 (2)
Ser289 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-10.7584
Phe291 (1)
Arg292 (4)
Ser294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
8 |
-12.0939
Trp282 (1)
Ser294 (1)
Arg292 (3)
Glu281 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-3.9584
Tyr 120 (1)
Ser294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | | 2 |
-10.3404
Glu281 (1)
Arg292 (3)
Trp282 (1)
Ser294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-8.6853
Trp282 (1)
Tyr68 (1)
Arg292 (3)
Ser 294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
9 |
-7.9259
Tyr68 (1)
Ser294 (1)
Arg292 (2)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
0.0528
Trp282 (2)
Ser294 (1)
Glu281 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | | 3 |
-7.7793
Trp282 (1)
Ser289 (1)
Arg292 (3)
Ser294 (3)
Glu281 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-7.7927
Trp282 (1)
Ser289 (1)
Arg292 (3)
Ser294 (3)
Glu281 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
10 |
-9.7289
Asp70 (1)
Tyr120 (1)
Ser294 (2)
Gly118 (1)
Tyr129 (1)
Glu198 (1) |
-10.7966
Arg292 (3)
Ser294 (2)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | | 4 |
-6.7650
Asp70 (1)
Phe291 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-9.7872
Tyr120 (2)
Ser294 (2)
Gly118 (1)
Tyr129 (1)
Glu198 (1) |
2PAM |
-5.9830
Trp282 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-5.4829
Trp282 (1)
Arg292 (3)
Ser294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | | 5 |
-7.4288
Asp70 (1)
Trp282 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-1.7457
Trp282 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
2PAM1 |
-1.6366
Trp282 (1)
Ser289 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
3.4180
Trp282 (1)
Arg292 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | |
|
|
|
|
| |
|
|
|
|
| |
|
|
|
|
(Continues) | |
|
|
|
|
| | (Continues) |
|
|
|
|
| | Oxime |
Anti |
Syn |
Oxime |
Anti |
Syn | | 6 |
-11.0145
Asp70 (2)
Ser294 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-8.1143
Trp282 (1)
Ser294 (3)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
2PAM2 |
-5.7507
Trp282 (1)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-5.8037
Trp282 (2)
Arg292 (2)
Ser294 (1)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) | | 7 |
-9.0249
Trp282 (2)
Ser294 (2)
Arg292 (2)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
-7.3808
Trp282 (1)
Ser294 (2)
Arg292 (3)
Gly118 (1)
Tyr120 (1)
Tyr129 (1)
Glu198 (1) |
|
|
|
| Oxime |
anti-anti |
syn-syn |
anti-syn | | TMB4 |
-10.4772
Ala200 (1)
Ser199 (1)
Gly118 (1)
Tyr120 (1)
Ser294 (1)
Glu281 (1)
Gly118 (1)
Tyr129 (1)
Glu198 (1) |
-5.0099
Tyr120 (1)
Ser294 (3)
Gly118 (1)
Tyr129 (1)
Glu198 (1) |
-9.2889
Gly118 (2)
Tyr120 (1)
Ser294 (3)
Gly118 (1)
Tyr129 (1)
Glu198 (1) | | Obidoxime |
12.3490
Ser294 (2)
His443 (1)
Tyr120 (1)
Glu198 (1)
Glu281 (1)
Ser199 (1)
Gly118 (1)
Tyr129 (1) |
-9.1658
Tyr68 (1)
Tyr120 (2)
Gly117 (1)
Gly118 (1)
Ser199 (1)
Tyr129 (1)
Glu198 (1) |
-15.5061
Ser294 (4)
Gly118 (1)
Tyr120 (3)
Ser199 (1)
Tyr129 (1)
Glu198 (1) |
Nowadays, despite the recent efforts at both
theoretical and experimental levels to elucidate the
reaction mechanism involved in the reactivation
process, some relevant facts, such as the influence of
the oxime's isomery, still need to be clarified. Fig. 7
displays the proposed reaction mechanism
considering 2PAM with syn and anti conformation.
2PAM was selected for the mechanism study due
to experimental evidence, which suggest that this
oxime could be used as a lead compound in order to
propose structures of new oximes, such as 2PAM1
and 2PAM2.
The syn and anti conformations present the
hydroxyl group close to the phosphate group of tabun,
with distance values around 3.14Å and 3.24Å
respectively (Fig. 8). For 2PAM1 and 2PAM2,
similar distances were obtained for amino acid
residues from the protein, favouring then, the reaction
process.
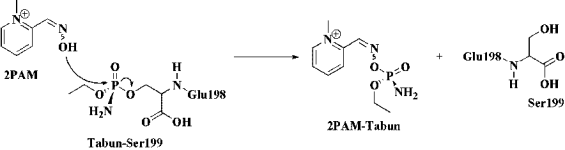
Fig. 7. Scheme of the reactivation mechanism of the acetylcholinesterase enzyme.
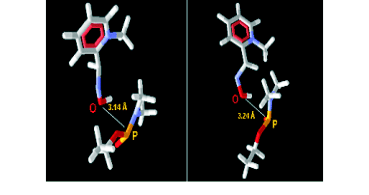
Fig. 8. Compound 2PAM in the anti and syn conformations, respectively.
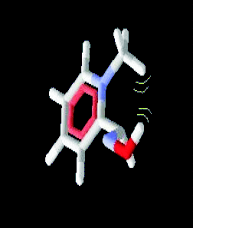
Fig. 9. Intermolecular steric effect in the compound
anti-2PAM.
From our theoretical calculations of the catalytic
mechanism, we obtained the relative activation
energy of the three isomers 2PAM, 2PAM1 and
2PAM2, in different conformations, syn and anti.
Theoretical data from Table 2 show that 2PAM and
2PAM2 in the syn conformation perform lower
activation energy values than the compounds in anti
conformations. This means that those conformations
revealed a lower energetic barrier for the reaction
pathway.
The lower thermodynamic stability of the 2PAM
isomers with anti conformation, can be rationalized
due to the lower number of hydrogen interactions
with the residues close to the active site (Trp282,
Arg292, Gly118, Tyr120, Tyr129, Glu198).
Regarding the compounds in the syn conformation
(Trp282, Arg292, Ser294, Gly118, Tyr120, Tyr129,
Glu198), we can observe a lower stability inside the
active site, suggesting a less stable transition state
(higher activation energy).
Table 2. Relative activation energy values of the studied oximes.
| Compounds |
E# (kcal mol-1) | | 2PAM syn |
31.98 | | 2PAM anti |
34.05 | | 2PAM1 syn |
10.30 | | 2PAM1 anti |
0.00 | | 2PAM2 syn |
23.88 | | 2PAM2 anti |
32.48 |
Turning now to the 2PAM1, our theoretical data
point out that the syn conformation presents a steric
hindrance between the aromatic hydrogen and the
hydroxyl group in the same compound. The same
scenario did not occur with 2PAM1 in the anti
conformation, leaving the hydroxyl group free to
interact with the phosphate group of the inhibitor.
This molecule, then, possesses significant internal
degrees of freedom, leading consequently to a lower
activation energy value for the compound in the anti
conformation.
Besides anti and syn conformations, the ortho,
meta and para substituent orientations in the
compounds, can also affect the transition state
stability. In this way, we noticed that the compounds
2PAM and 2PAM2 have a methyl group in
ortho-orientation, generating a higher steric hindrance
with the hydroxyl group in the anti conformation,
resulting in a higher activation energy value in
relation to all other compounds, as reported in Fig. 9
and Table 2.
CONCLUSION
From the results obtained and discussed here it is
possible to conclude that: 1) oximes derived from
pyridine imidazol bicycled systems are worth
synthesizing and testing in vitro as HssAChE
reactivators and are expected to present similar
experimental results as TMB-4; 2) the position of the
oxime group as substituent, in the six or the five
membered ring, on these molecules seems not to have
a determinant influence on their affinities for the
HssAChE active site; 3) meta-2PAM should be
considered further as an HssAChE reactivator in
experimental studies and 4) it is important, also, to
consider a deeper investigation of the influence of the
anti isomers in experimental studies of oximes as
HssAChE reactivators.
ACKNOWLEDGMENTS
The authors wish to thank the Brazilian financial
agencies CNPq, FAPERJ, FAPEMIG and
CAPES/PRODEFESA for financial support and the
Military Institute of Engineering for the physical
infrastructure and working space. This work was also
supported by Ministry of Defense (Czech Republic) -
FVZUO0000604 (KK).
REFERENCES
Bartosova L, Kuca K, Jun D, Kunesova G. Bispyridinium oximes as antidotal treatment of cyclosarin poisoning - in vitro and in vivo testing. Internat J Tox. 24: 399-402, 2005.
[CrossRef]
Bay E, Krop S, Yates LF. Chemotherapeutic effectiveness of 1,1’-trimethylene bis (4-formylpyridinium bromide) di-oxime (TMB-4) in experimental anticholinesterase poisoning. Proc Soc Exp Biol Med. 98: 107-110, 1958.
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat T N, Weissig H, Shindyalov IN, Bourne PE. The protein data bank. Nucleic Acids Res. 28: 235-242, 2000.
[CrossRef]
Bernstein FC, Koetzle TF, Williams GJ, Meyer EE, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein data bank. A computer-based archival file for macromolecular structures. J Mol Biol. 112: 535- 542, 1977.
[CrossRef]
Castro AT, Figueroa-Villar JD. Molecular structure, conformational analysis and charge distribution of pralidoxime: Ab initio and DFT studies. Int J Quantum Chem. 89: 135-146, 2002.
[CrossRef]
Delfino RT, Figueroa-Villar JD. Nucleophilic reactivation of sarin-inhibited acetylcholinesterase: A molecular modeling study, J Phys Chem B. 113: 8402-8411, 2009.
[CrossRef]
Delfino RT, Ribeiro TS, Figueroa-Villar JD. Organophosphorus compounds as chemical warfare agents: a review. J Braz Chem Soc. 20: 407-428, 2009.
[CrossRef]
Ekstrom F J, Astot C, Pang YP. Novel nerve-agent antidote design based on crystallographic and mass spectrometric analyses of tabun-conjugated acetylcholinesterase in complex with antidotes, Clin Pharmacol Ther. 82: 282-293, 2007.
[CrossRef]
Frisch MJ, Trucks GW, Schlegel HB, Scuseria ES, Pople JA, Gaussian 98 (Revision A.11), Gaussian: Pittsburgh (2001).
Goncalves AS, Franca TCC, Wilter A, Figueroa-Villar JD. Molecular dynamics of the interaction of pralidoxime and deazapralidoxime with acetylcholinesterase inhibited by the neurotoxic agent tabun. J Braz Chem Soc. 17: 968-975, 2006.
[CrossRef]
Goncalves AS, França TCC, Figueroa-Villar JD, Pascutti PG. Conformational analysis of toxogonine, TMB-4 and HI-6 using PM6 and RM1 methods. J Braz Chem Soc. 21: 179-184, 2010.
[CrossRef]
Hehre WJ, Deppmeier BJ, Klunzinger PE, PC SPARTAN Pro, Wavefunction Inc.: Irvine, California (1999).
Kassa J, Kuca K, Bartosova L, Kunesova G. The development of new structural analogues of oximes for the antidotal treatment of poisoning by nerve agents and the comparison of their reactivating and therapeutic efficacy with currently available oximes. Curr Org Chem. 11: 267-283, 2007.
[CrossRef]
Kryger G, Harel M, Giles K, Toker L, Velan B, Lazar A, Kronman C, Barak D, Ariel N, Silman I, Shafferman A, Sussman JL. Structures of recombinant native and E202Q mutant human acetylcholinesterase complexed with the snake-venom toxin fasciculin-II. Acta Crystallogr. 56: 1385-1394, 2000.
Ramalho TC, Taft CA. Thermal and solvent effects on the NMR and UV parameters of some bioreductive drugs. J Chem Physics. 123: 54319-54328, 2005.
[CrossRef]
Ramalho TC, Franca TCC, Renno MN, Guimaraes AP, da Cunha EFF, Kuca K. Development of new acetylcholinesterase reactivators: Molecular modeling versus in vitro data. Chem Biol Inter. 185: 73-77, 2010.
[CrossRef]
Smirnova OI, E Gurina IL, Zhigalova V, Arestova LS. Toxicity and tolerance of toxogonin, Farmakol Toksikol. 38: 467-470, 1975.
Thomsen R, Christensen MH. MolDock: A new technique for high-accuracy molecular docking. J Med Chem. 49: 3315-3321, 2006.
[CrossRef]
Worek F, Aurbek N, Koller M, Becker C, Eyer P, Thiermann H. Kinetic analysis of reactivation and aging of human acetylcholinesterase inhibited by different phosphoramidates. Biochem Pharmacol. 73: 1807-1817, 2007.
[CrossRef]
|
BACK
|